3A6F
 
 | | W174F mutant creatininase, Type II | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6G
 
 | | W154F mutant creatininase | | Descriptor: | Creatinine amidohydrolase, MANGANESE (II) ION, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
1UCY
 
 | |
1UI6
 
 | |
1UI5
 
 | |
3ADE
 
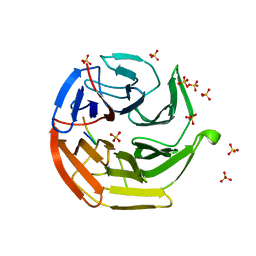 | | Crystal Structure of Keap1 in Complex with Sequestosome-1/p62 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Sequestosome-1 | | Authors: | Kurokawa, H, Yamamoto, M. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1
Nat.Cell Biol., 12, 2010
|
|
3AN3
 
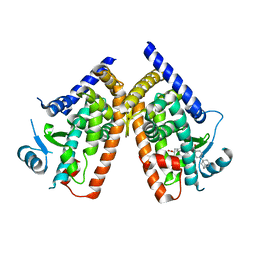 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist MO3S | | Descriptor: | (2S)-2-benzyl-3-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}phenyl)propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Waku, T, Miyachi, H, Morikawa, K. | | Deposit date: | 2010-08-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Phenylpropanoic Acid-Type PPAR gamma-Selective Agonists: Discovery of Reversed Stereochemistry-Activity Relationship
J.Med.Chem., 54, 2011
|
|
3AN4
 
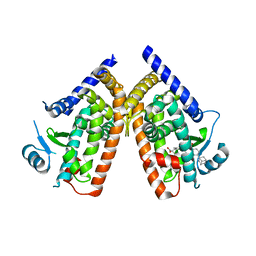 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist MO4R | | Descriptor: | (2R)-2-benzyl-3-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}phenyl)propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Ohashi, M, Waku, T, Miyachi, H, Morikawa, K. | | Deposit date: | 2010-08-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Phenylpropanoic Acid-Type PPAR gamma-Selective Agonists: Discovery of Reversed Stereochemistry-Activity Relationship
J.Med.Chem., 54, 2011
|
|
3WPW
 
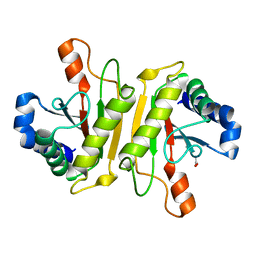 | | Structure of PomBc5, a periplasmic fragment of PomB from Vibrio | | Descriptor: | ACETATE ION, PomB | | Authors: | Takao, M, Sakuma, M, Zhu, S, Homma, M, Kojima, S, Imada, K. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational change in the periplasmic region of the flagellar stator coupled with the assembly around the rotor
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
3WPX
 
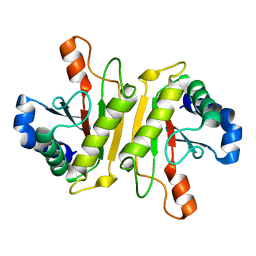 | | Structure of PomBc4, a periplasmic fragment of PomB from Vibrio alginolyticus | | Descriptor: | PomB | | Authors: | Takao, M, Sakuma, M, Zhu, S, Homma, M, Kojima, S, Imada, K. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational change in the periplasmic region of the flagellar stator coupled with the assembly around the rotor
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
7VKF
 
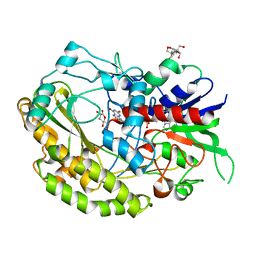 | | Reduced enzyme of FAD-dpendent Glucose Dehydrogenase complex with D-glucono-1,5-lactone at pH8.5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-glucono-1,5-lactone, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y, Nishiya, Y, Ito, K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
7VKD
 
 | |
2DCE
 
 | | Solution structure of the SWIRM domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Tochio, N, Umehara, T, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-06 | | Release date: | 2006-07-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2CU7
 
 | | Solution structure of the SANT domain of human KIAA1915 protein | | Descriptor: | KIAA1915 protein | | Authors: | Yoneyama, M, Umehara, T, Saito, K, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-25 | | Release date: | 2005-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
2CUJ
 
 | | Solution structure of SWIRM domain of mouse transcriptional adaptor 2-like | | Descriptor: | transcriptional adaptor 2-like | | Authors: | Yoneyama, M, Umehara, T, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Differences of SWIRM Domain Subtypes
J.Mol.Biol., 369, 2007
|
|
1MKX
 
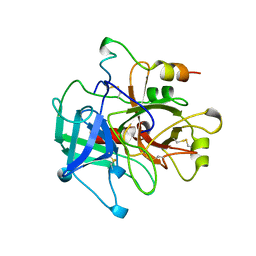 | |
2DDH
 
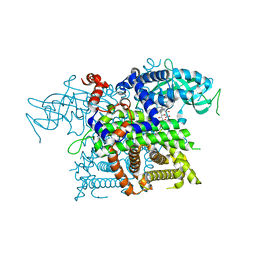 | | Crystal Structure of Acyl-CoA oxidase complexed with 3-OH-dodecanoate | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Acyl-CoA oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Keiji, T, Nakajima, Y, Miyahara, I, Hirotsu, K. | | Deposit date: | 2006-01-29 | | Release date: | 2006-03-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Three-Dimensional Structure of Rat-Liver Acyl-CoA Oxidase in Complex with a Fatty Acid: Insights into Substrate-Recognition and Reactivity toward Molecular Oxygen.
J.Biochem.(Tokyo), 139, 2006
|
|
1YCP
 
 | |
2D7T
 
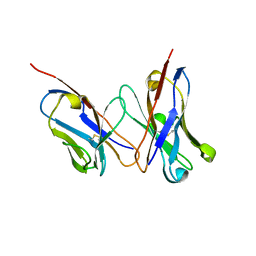 | |
5XRZ
 
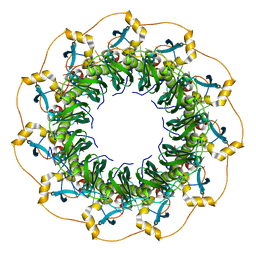 | | Structure of a ssDNA bound to the inner DNA binding site of RAD52 | | Descriptor: | DNA repair protein RAD52 homolog, POTASSIUM ION, ssDNA (40-MER) | | Authors: | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | Deposit date: | 2017-06-11 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
5XQV
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20L mutant (NFE6, AFP), P21 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XR0
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20T mutant (NFE6, AFP), P21 form | | Descriptor: | Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3A1R
 
 | |
