2O4A
 
 | |
2O49
 
 | |
3OTJ
 
 | | A Crystal Structure of Trypsin Complexed with BPTI (Bovine Pancreatic Trypsin Inhibitor) by X-ray/Neutron Joint Refinement | | Descriptor: | CALCIUM ION, Cationic trypsin, Pancreatic trypsin inhibitor, ... | | Authors: | Kawamura, K, Yamada, T, Kurihara, K, Tamada, T, Kuroki, R, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2010-09-12 | | Release date: | 2011-01-26 | | Last modified: | 2017-11-08 | | Method: | NEUTRON DIFFRACTION (2.15 Å), X-RAY DIFFRACTION | | Cite: | X-ray and neutron protein crystallographic analysis of the trypsin-BPTI complex.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2CVH
 
 | | Crystal structure of the RadB recombinase | | Descriptor: | DNA repair and recombination protein radB | | Authors: | Akiba, T, Ishii, N, Rashid, N, Morikawa, M, Imanaka, T, Harata, K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-08-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of RadB recombinase from a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1: an implication for the formation of a near-7-fold helical assembly
Nucleic Acids Res., 33, 2005
|
|
2CVF
 
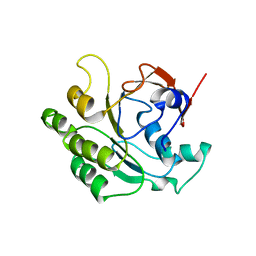 | | Crystal structure of the RadB recombinase | | Descriptor: | DNA repair and recombination protein radB | | Authors: | Akiba, T, Ishii, N, Rashid, N, Morikawa, M, Imanaka, T, Harata, K. | | Deposit date: | 2005-06-03 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of RadB recombinase from a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1: an implication for the formation of a near-7-fold helical assembly
Nucleic Acids Res., 33, 2005
|
|
2DHT
 
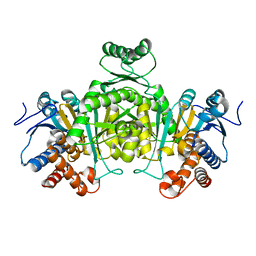 | |
2EFC
 
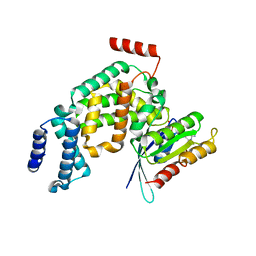 | | Ara7-GDP/AtVps9a | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Similarity to vacuolar protein sorting-associated protein VPS9, Small GTP-binding protein-like | | Authors: | Uejima, T, Ihara, K, Goh, T, Ito, E, Sunada, M, Ueda, T, Nakano, A, Wakatsuki, S. | | Deposit date: | 2007-02-22 | | Release date: | 2008-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | GDP-bound and nucleotide-free intermediates of the guanine nucleotide exchange in the Rab5/Vps9 system
J.Biol.Chem., 285, 2010
|
|
3CU0
 
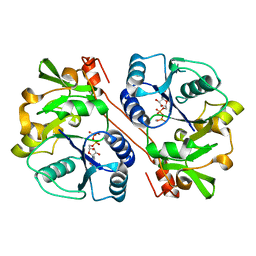 | | human beta 1,3-glucuronyltransferase I (GlcAT-I) in complex with UDP and GAL-GAL(6-SO4)-XYL(2-PO4)-O-SER | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 3, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Tone, Y, Pedersen, L.C, Yamamoto, T, Kitagawa, H, Nishihara-Shimizu, J, Tamura, J, Negishi, M, Sugahara, K. | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-o-phosphorylation of xylose and 6-o-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis.
J.Biol.Chem., 283, 2008
|
|
3WMY
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WMZ
 
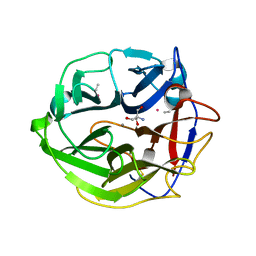 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase ethylmercury derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WN2
 
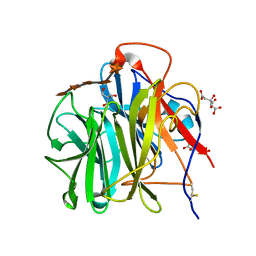 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylohexaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WA3
 
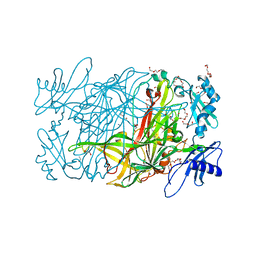 | | Crystal structure of copper amine oxidase from arthrobacter globiformis in N2 condition | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WJL
 
 | | Crystal structure of IIb selective Fc variant, Fc(V12), in complex with FcgRIIb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-c, ... | | Authors: | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
3WA2
 
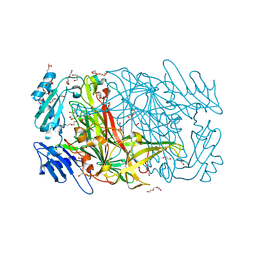 | | High resolution crystal structure of copper amine oxidase from arthrobacter globiformis | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murakawa, T, Hayashi, H, Sunami, T, Kurihara, K, Tamada, T, Kuroki, R, Suzuki, M, Tanizawa, K, Okajima, T. | | Deposit date: | 2013-04-22 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-resolution crystal structure of copper amine oxidase from Arthrobacter globiformis: assignment of bound diatomic molecules as O2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GPG
 
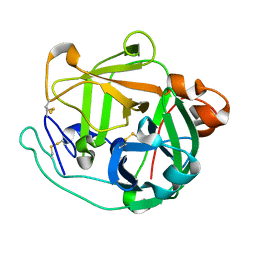 | | X/N joint refinement of Achromobacter Lyticus Protease I free form at pD8.0 | | Descriptor: | Protease 1 | | Authors: | Ohnishi, Y, Yamada, T, Kurihara, K, Tanaka, I, Sakiyama, F, Masaki, T, Niimura, N. | | Deposit date: | 2012-08-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | NEUTRON DIFFRACTION (1.895 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of Achromobacter protease I at pD 8.0: protonation states and hydration structure in the free-form.
Biochim.Biophys.Acta, 1834, 2013
|
|
3WJJ
 
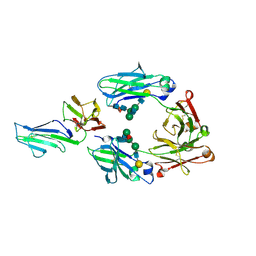 | | Crystal structure of IIb selective Fc variant, Fc(P238D), in complex with FcgRIIb | | Descriptor: | Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-b, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
2Z97
 
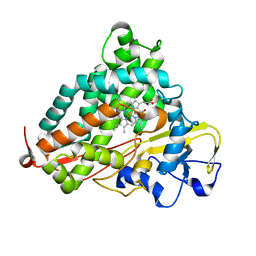 | | Crystal Structure of Ferric Cytochrome P450cam Reconstituted with 7-Methyl-7-depropionated Hemin | | Descriptor: | 7-METHYL-7-DEPROPIONATEHEMIN, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Hayashi, T, Harada, K, Sakurai, K, Hirota, S, Shimada, H. | | Deposit date: | 2007-09-18 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A role of the heme-7-propionate side chain in cytochrome P450cam as a gate for regulating the access of water molecules to the substrate-binding site
J.Am.Chem.Soc., 131, 2009
|
|
3WN1
 
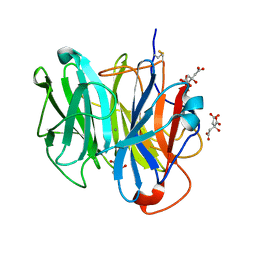 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WN0
 
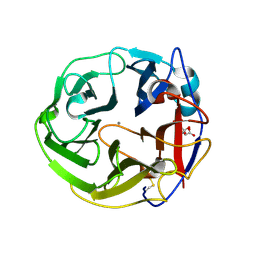 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with L-arabinose | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
4FW9
 
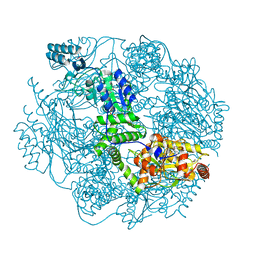 | | Crystal structure of the Lon-like protease MtaLonC | | Descriptor: | PHOSPHATE ION, TTC1975 peptidase | | Authors: | Chang, C.I, Ihara, K, Kuo, C.I, Huang, K.F, Wakatsuki, S. | | Deposit date: | 2012-06-30 | | Release date: | 2013-06-26 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of an ATP-independent Lon-like protease and its complexes with covalent inhibitors
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2IEY
 
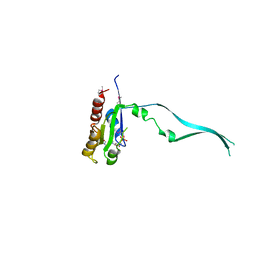 | | Crystal Structure of mouse Rab27b bound to GDP in hexagonal space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IF0
 
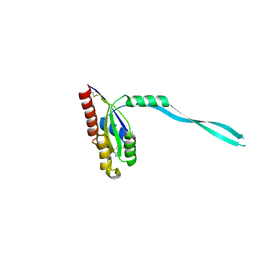 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2IEZ
 
 | | Crystal Structure of mouse Rab27b bound to GDP in monoclinic space group | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3FHP
 
 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | Descriptor: | Insulin, ZINC ION | | Authors: | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3VOR
 
 | | Crystal Structure Analysis of the CofA | | Descriptor: | CFA/III pilin | | Authors: | Fukakusa, S, Kawahara, K, Nakamura, S, Iwasita, T, Baba, S, Nishimura, M, Kobayashi, Y, Honda, T, Iida, T, Taniguchi, T, Ohkubo, T. | | Deposit date: | 2012-02-06 | | Release date: | 2012-09-26 | | Last modified: | 2013-07-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structure of the CFA/III major pilin subunit CofA from human enterotoxigenic Escherichia coli determined at 0.90 A resolution by sulfur-SAD phasing
Acta Crystallogr.,Sect.D, 68, 2012
|
|
