4PZ0
 
 | | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2) | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of a solute binding protein from Bacillus anthracis str. Ames in complex with quorum-sensing signal autoinducer-2 (AI-2).
To be Published
|
|
4M0C
 
 | | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | The crystal structure of a FMN-dependent NADH-azoreductase from Bacillus anthracis str. Ames Ancestor in complex with FMN.
To be Published
|
|
4M0G
 
 | | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor. | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Tan, K, Zhou, M, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor.
To be Published
|
|
4R7Q
 
 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4MAM
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP
To be Published
|
|
4M9U
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4RN7
 
 | | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | The crystal structure of N-acetylmuramoyl-L-alanine amidase from Clostridium difficile 630
To be Published
|
|
4MA0
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP
To be Published
|
|
4M9D
 
 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
4MA5
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP. | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP.
To be Published
|
|
4S1N
 
 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Streptococcus pneumoniae TIGR4
To be Published
|
|
4NEG
 
 | | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | FORMIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhang, R, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The crystal structure of tryptophan synthase subunit beta from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
4NPB
 
 | | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92 | | Descriptor: | PHOSPHATE ION, Protein disulfide isomerase II, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | The crystal structure of thiol:disulfide interchange protein DsbC from Yersinia pestis CO92
To be Published
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-11 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4YYF
 
 | | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155 | | Descriptor: | ACETATE ION, Beta-N-acetylhexosaminidase, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155
To Be Published
|
|
7JFQ
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
5CD2
 
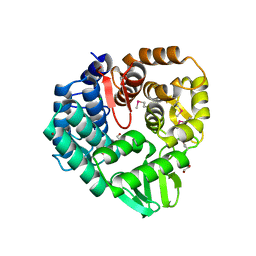 | | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114 | | Descriptor: | CHLORIDE ION, Endo-1,4-D-glucanase, GLYCEROL, ... | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-02 | | Release date: | 2015-07-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of endo-1,4-D-glucanase from Vibrio fischeri ES114
To Be Published
|
|
5C0P
 
 | | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endo-arabinase, ... | | Authors: | Tan, K, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482
To Be Published
|
|
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
5D5H
 
 | | Crystal structure of Mycobacterium tuberculosis Topoisomerase I | | Descriptor: | ACETATE ION, DNA topoisomerase 1, GLYCEROL, ... | | Authors: | Tan, K, Cheng, B, Tse-Dinh, Y.C. | | Deposit date: | 2015-08-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold.
J.Mol.Biol., 428, 2016
|
|
4DQD
 
 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4DQ0
 
 | |
4DZR
 
 | | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The crystal structure of protein-(glutamine-N5) methyltransferase (release factor-specific) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
4EDH
 
 | | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tan, K, Joachimiak, G, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The crystal structure of thymidylate kinase from Pseudomonas aeruginosa PAO1 in complex with ADP,TMP and Mg.
To be Published
|
|
4ER9
 
 | | Crystal structure of cytochrome b562 from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Jedrzejczak, R, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Crystal structure of cytochrome b562 from Salmonella enterica subsp. enterica serovar Typhimurium str. 14028S
To be Published
|
|
