2BR8
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with an alpha-conotoxin PnIA variant | | Descriptor: | ALPHA-CONOTOXIN PNIA, SOLUBLE ACETYLCHOLINE RECEPTOR, SULFATE ION | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, van Nierop, P, van Elk, R, van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha-Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
2CDP
 
 | | Structure of a CBM6 in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-26 | | Release date: | 2006-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
2BJ0
 
 | | Crystal Structure of AChBP from Bulinus truncatus revals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Klaassen, R.V, Van Rossum-Fikkert, S.E, Van Elk, R, Van Nierop, P, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-01-27 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Acetylcholine-Binding Protein from Bulinus Truncatus Reveals the Conserved Structural Scaffold and Sites of Variation in Nicotinic Acetylcholine Receptors.
J.Biol.Chem., 280, 2005
|
|
2EPE
 
 | | Crystal structure analysis of Hen egg white lysozyme grown by capillary method | | Descriptor: | Lysozyme C | | Authors: | Naresh, M.D, Subramanian, V, Jaimohan, S.M, Rajaram, A, Arumugam, V, Usha, R, Mandal, A.B. | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of Hen egg white lysozyme grown by capillary method
To be published
|
|
2C3G
 
 | | Structure of CBM26 from Bacillus halodurans amylase | | Descriptor: | ALPHA-AMYLASE G-6, CADMIUM ION | | Authors: | Boraston, A.B, Healey, M, Klassen, J, Ficko-Blean, E, Lammerts Van Bueren, A, Law, V. | | Deposit date: | 2005-10-07 | | Release date: | 2005-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural and Functional Analysis of Alpha-Glucan Recognition by Family 25 and 26 Carbohydrate-Binding Modules Reveals a Conserved Mode of Starch Recognition
J.Biol.Chem., 281, 2006
|
|
2CNJ
 
 | | NMR studies on the interaction of Insulin-Growth Factor II (IGF-II) with IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Williams, C, Prince, S, Zaccheo, O, Hassan, A.B, Crosby, J, Crump, M. | | Deposit date: | 2006-05-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights Into the Interaction of Insulin-Like Growth Factor 2 with Igf2R Domain 11.
Structure, 15, 2007
|
|
2C9T
 
 | | Crystal Structure Of Acetylcholine Binding Protein (AChBP) From Aplysia Californica In Complex With alpha-Conotoxin ImI | | Descriptor: | ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Hogg, R.C, Celie, P.H, Bertrand, D, Tsetlin, V, Smit, A.B, Sixma, T.K. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Determinants of Selective {Alpha}-Conotoxin Binding to a Nicotinic Acetylcholine Receptor Homolog Achbp.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CDO
 
 | | structure of agarase carbohydrate binding module in complex with neoagarohexaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, BETA-AGARASE 1, ... | | Authors: | Henshaw, J, Horne, A, Van Bueren, A.L, Money, V.A, Bolam, D.N, Czjzek, M, Weiner, R.M, Hutcheson, S.W, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2006-01-25 | | Release date: | 2006-02-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 6 Carbohydrate Binding Modules in Beta-Agarases Display Exquisite Selectivity for the Non- Reducing Termini of Agarose Chains.
J.Biol.Chem., 281, 2006
|
|
2C3W
 
 | | Structure of CBM25 from Bacillus halodurans amylase in complex with maltotetraose | | Descriptor: | ALPHA-AMYLASE G-6, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Boraston, A.B, Healey, M, Klassen, J, Ficko-Blean, E, Lammerts van Bueren, A, Law, V. | | Deposit date: | 2005-10-12 | | Release date: | 2005-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A Structural and Functional Analysis of Alpha-Glucan Recognition by Family 25 and 26 Carbohydrate-Binding Modules Reveals a Conserved Mode of Starch Recognition
J.Biol.Chem., 281, 2006
|
|
2BW3
 
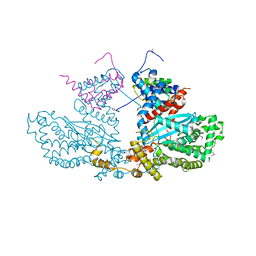 | | Three-dimensional structure of the Hermes DNA transposase | | Descriptor: | TRANSPOSASE | | Authors: | Hickman, A.B, Perez, Z.N, Zhou, L, Musingarimi, P, Ghirlando, R, Hinshaw, J.E, Craig, N.L, Dyda, F. | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of a Eukaryotic DNA Transposase
Nat.Struct.Mol.Biol., 12, 2005
|
|
2C3H
 
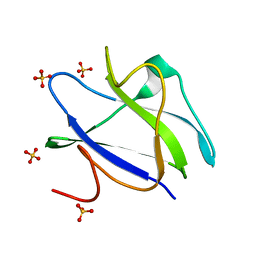 | | Structure of CBM26 from Bacillus halodurans amylase in complex with maltose | | Descriptor: | ALPHA-AMYLASE G-6, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Boraston, A.B, Healey, M, Klassen, J, Ficko-Blean, E, Lammerts van Bueren, A, Law, V. | | Deposit date: | 2005-10-07 | | Release date: | 2005-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A Structural and Functional Analysis of Alpha-Glucan Recognition by Family 25 and 26 Carbohydrate-Binding Modules Reveals a Conserved Mode of Starch Recognition
J.Biol.Chem., 281, 2006
|
|
2CGR
 
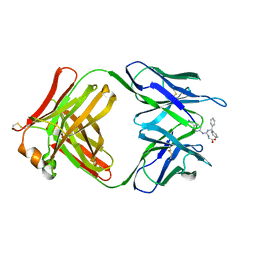 | |
2BB9
 
 | | Structure of HIV1 protease and AKC4p_133a complex. | | Descriptor: | 2-ETHOXYETHYL (1S,2S)-3-{(2S)-4-[(3AS,8S,8AR)-2-OXO-3,3A,8,8A-TETRAHYDRO-2H-INDENO[1,2-D][1,3]OXAZOL-8-YL]-2-BENZYL-3-OXO-2,3-DIHYDRO-1H-PYRROL-2-YL}-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Smith III, A.B, Charnley, A.K, Kuo, L.C, Munshi, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2CG4
 
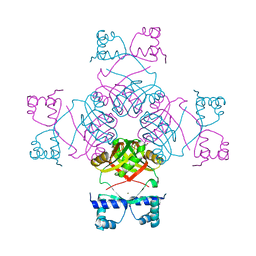 | | Structure of E.coli AsnC | | Descriptor: | ASPARAGINE, MAGNESIUM ION, REGULATORY PROTEIN ASNC | | Authors: | Thaw, P, Sedelnikova, S.E, Muranova, T, Wiese, S, Ayora, S, Alonso, J.C, Brinkman, A.B, Akerboom, J, van der Oost, J, Rafferty, J.B. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into Gene Transcriptional Regulation and Effector Binding by the Lrp/Asnc Family.
Nucleic Acids Res., 34, 2006
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
1LHZ
 
 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 293K | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | Deposit date: | 2002-04-17 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
1LGV
 
 | | Structure of a Human Bence-Jones Dimer Crystallized in U.S. Space Shuttle Mission STS-95: 100K | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Terzyan, S.S, DeWitt, C.R, Ramsland, P.A, Bourne, P.C, Edmundson, A.B. | | Deposit date: | 2002-04-16 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of a human Bence-Jones dimer
crystallized on Earth and aboard US Space Shuttle Mission STS-95
J.MOL.RECOG., 16, 2003
|
|
1I9B
 
 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, CALCIUM ION | | Authors: | Brejc, K, van Dijk, W.J, Klaassen, R, Schuurmans, M, van der Oost, J, Smit, A.B, Sixma, T.K. | | Deposit date: | 2001-03-18 | | Release date: | 2001-05-16 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors.
Nature, 411, 2001
|
|
3A59
 
 | |
2YA0
 
 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | Authors: | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
2YGL
 
 | | The X-ray crystal structure of tandem CBM51 modules of Sp3GH98, the family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | Descriptor: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
2Y7Y
 
 | | APLYSIA CALIFORNICA ACHBP IN APO STATE | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, De Esch, I.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X-Ray Analysis.
J.Med.Chem., 52, 2009
|
|
2YB8
 
 | | Crystal structure of Nurf55 in complex with Su(z)12 | | Descriptor: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | Authors: | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
3B86
 
 | |
