1DFN
 
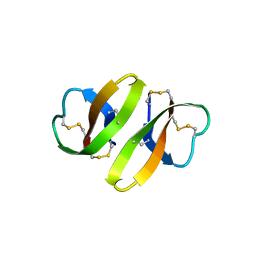 | | CRYSTAL STRUCTURE OF DEFENSIN HNP-3, AN AMPHIPHILIC DIMER: MECHANISMS OF MEMBRANE PERMEABILIZATION | | Descriptor: | DEFENSIN HNP-3 | | Authors: | Hill, C.P, Yee, J, Selsted, M.E, Eisenberg, D. | | Deposit date: | 1991-01-18 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization.
Science, 251, 1991
|
|
6SKL
 
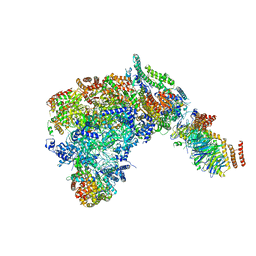 | | Cryo-EM structure of the CMG Fork Protection Complex at a replication fork - Conformation 1 | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA fork, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
6SKO
 
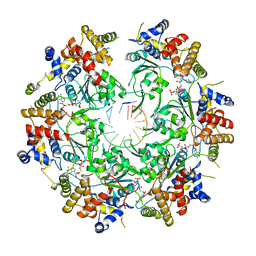 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
3BDF
 
 | |
3BDG
 
 | |
3BDH
 
 | |
