4TTO
 
 | |
4TTM
 
 | | Racemic structure of kalata B1 (kB1) | | Descriptor: | D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9001 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4TTL
 
 | |
4TTK
 
 | |
4TTN
 
 | | Quasi-racemic structure of [G6A]kalata B1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-kalata B1, Kalata-B1 | | Authors: | Wang, C.K, King, G.J, Craik, D.J. | | Deposit date: | 2014-06-22 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2507 Å) | | Cite: | Racemic and Quasi-Racemic X-ray Structures of Cyclic Disulfide-Rich Peptide Drug Scaffolds.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5INZ
 
 | | Racemic structure of baboon theta defensin-2 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Theta defensin-2, ... | | Authors: | Wang, C.K, King, G.J, Conibear, A.C, Ramos, M.C, Craik, D.J. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Mirror Images of Antimicrobial Peptides Provide Reflections on Their Functions and Amyloidogenic Properties.
J.Am.Chem.Soc., 138, 2016
|
|
2KHB
 
 | |
2K7G
 
 | | Solution Structure of varv F | | Descriptor: | Varv peptide F | | Authors: | Wang, C.K. | | Deposit date: | 2008-08-10 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Combined X-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold
J.Biol.Chem., 284, 2009
|
|
2KCG
 
 | | Solution structure of cycloviolacin O2 | | Descriptor: | Cycloviolacin-O2 | | Authors: | Wang, C.K. | | Deposit date: | 2008-12-22 | | Release date: | 2009-07-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes.
Biophys.J., 97, 2009
|
|
2KCH
 
 | | Solution structure of micelle-bound kalata B2 | | Descriptor: | Kalata-B2 | | Authors: | Wang, C.K. | | Deposit date: | 2008-12-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes.
Biophys.J., 97, 2009
|
|
2KVX
 
 | | Solution structure of kalata B12 | | Descriptor: | Kalata-B12 | | Authors: | Wang, C.K. | | Deposit date: | 2010-03-29 | | Release date: | 2011-03-09 | | Last modified: | 2016-06-01 | | Method: | SOLUTION NMR | | Cite: | The role of conserved Glu residue on cyclotide stability and activity: a structural and functional study of kalata B12, a naturally occurring Glu to Asp mutant.
Biochemistry, 50, 2011
|
|
6DL1
 
 | | Racemic structure of jatrophidin, an orbitide from Jatropha curcas | | Descriptor: | jatrophidin | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.029 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6DL0
 
 | | Crystal structure of pohlianin C, an orbitide from Jatropha pohliana | | Descriptor: | pohlianin C | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6DKZ
 
 | | Racemic structure of ribifolin, an orbitide from Jatropha ribifolia | | Descriptor: | ribifolin | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-14 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6DKY
 
 | | Crystal structure of ribifolin, an orbitide from Jatropha ribifolia | | Descriptor: | ILE-LEU-GLY-SER-ILE-ILE-LEU-GLY | | Authors: | Wang, C.K, Ramalho, S.D, King, G.J, Craik, D.J. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
8GCR
 
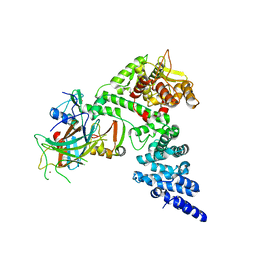 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
7RIJ
 
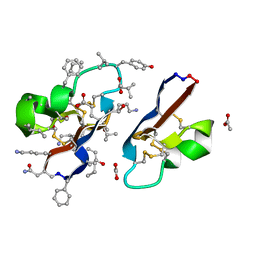 | | [I11G]hyen D | | Descriptor: | ACETATE ION, Cyclotide hyen-D, D-[I11L]hyen D | | Authors: | Du, Q, Huang, Y.H, Wang, C.K, Craik, D.J. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Enabling efficient folding and high-resolution crystallographic analysis of bracelet cyclotides
Molecules, 26(18), 2021
|
|
7RIH
 
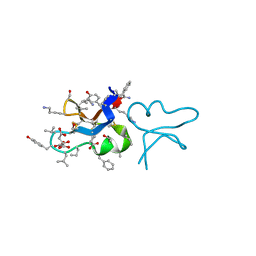 | | hyen D | | Descriptor: | CITRATE ANION, Cyclotide hyen-D, D-[I11L]hyen D | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enabling efficient folding and high-resolution crystallographic analysis of bracelet cyclotides
Molecules, 26(18), 2021
|
|
7RII
 
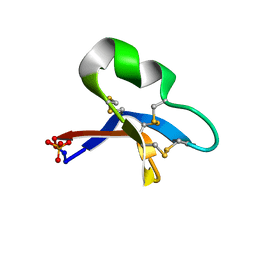 | | [I11L]hyen D crystal structure | | Descriptor: | Cyclotide hyen-D, PHOSPHATE ION | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Enabling efficient folding and high-resolution crystallographic analysis of bracelet cyclotides
Molecules, 26(18), 2021
|
|
4LRV
 
 | | Crystal structure of DndE from Escherichia coli B7A involved in DNA phosphorothioation modification | | Descriptor: | DNA sulfur modification protein DndE | | Authors: | Hu, W, Wang, C.K, Liang, J.D, Zhang, T.L, Yang, M, Hu, Z.P, Wang, Z.J, Lan, W.X, Wu, H.M, Ding, J.P, Wu, G, Deng, Z.X, Cao, C. | | Deposit date: | 2013-07-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification
Cell Res., 22, 2012
|
|
6U7X
 
 | | NMR solution structure of triazole bridged plasmin inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Wang, C.K, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7RN3
 
 | | hyen D solution structure | | Descriptor: | Cyclotide hyen-D | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-29 | | Release date: | 2022-03-02 | | Last modified: | 2022-12-07 | | Method: | SOLUTION NMR | | Cite: | Mutagenesis of bracelet cyclotide hyen D reveals functionally and structurally critical residues for membrane binding and cytotoxicity.
J.Biol.Chem., 298, 2022
|
|
3T85
 
 | |
3T84
 
 | |
3T82
 
 | |
