6E61
 
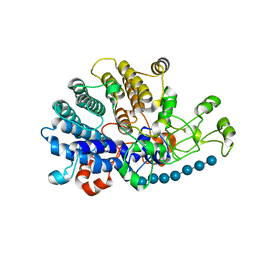 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-A in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-23 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6E9B
 
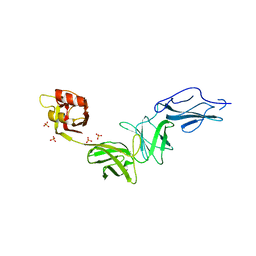 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-B in complex with mixed-linkage heptasaccharide | | Descriptor: | Mixed-linkage glucan utilization locus (MLGUL) SGBP-B, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6E60
 
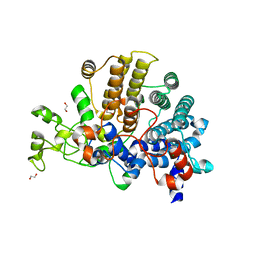 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-A | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, mixed-linkage glucan utilization locus (MLGUL) SGBP-B | | Authors: | Tamura, K, Gardill, B.R, Brumer, H, Van Petegem, F. | | Deposit date: | 2018-07-23 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
1NCJ
 
 | | N-CADHERIN, TWO-DOMAIN FRAGMENT | | Descriptor: | CALCIUM ION, PROTEIN (N-CADHERIN), URANYL (VI) ION | | Authors: | Tamura, K, Shan, W.-S, Hendrickson, W.A, Colman, D.R, Shapiro, L. | | Deposit date: | 1999-02-02 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-function analysis of cell adhesion by neural (N-) cadherin.
Neuron, 20, 1998
|
|
7KV6
 
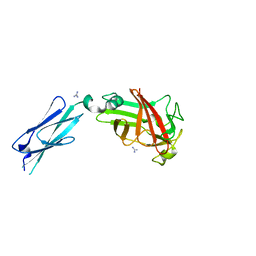 | |
7KWB
 
 | |
7KV7
 
 | | Surface glycan-binding protein B from Bacteroides fluxus in complex with laminaritriose | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, LYSINE, ... | | Authors: | Tamura, K, Brumer, H, Van Petegem, F. | | Deposit date: | 2020-11-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota.
J.Biol.Chem., 296, 2021
|
|
7KV5
 
 | |
7KWC
 
 | |
7KV1
 
 | | Surface glycan-binding protein A from Bacteroides uniformis | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT HEXAMMINE(III), ... | | Authors: | Tamura, K, Brumer, H, Van Petegem, F. | | Deposit date: | 2020-11-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota.
J.Biol.Chem., 296, 2021
|
|
7KV2
 
 | | Surface glycan-binding protein A from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RagB/SusD family nutrient uptake outer membrane protein, ... | | Authors: | Tamura, K, Brumer, H, Van Petegem, F. | | Deposit date: | 2020-11-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota.
J.Biol.Chem., 296, 2021
|
|
7KV3
 
 | |
7KV4
 
 | |
6PAL
 
 | | Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus | | Descriptor: | ACETATE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tamura, K, Brumer, H, van Petegem, F. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Synergy between Cell Surface Glycosidases and Glycan-Binding Proteins Dictates the Utilization of Specific Beta(1,3)-Glucans by Human GutBacteroides.
Mbio, 11, 2020
|
|
5NBO
 
 | | Bacteroides ovatus mixed linkage glucan PUL (MLGUL) GH16 | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 16 | | Authors: | Hemsworth, G.R, Tamura, K, Dejean, G, Rogers, T.E, Pudlo, N.A, Urs, K, Jain, N, Martens, E.C, Brumer, H, Davies, G.J. | | Deposit date: | 2017-03-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism by which Prominent Human Gut Bacteroidetes Utilize Mixed-Linkage Beta-Glucans, Major Health-Promoting Cereal Polysaccharides.
Cell Rep, 21, 2017
|
|
5NBP
 
 | | Bacteroides ovatus mixed linkage glucan PUL (MLGUL) GH16 in complex with G4G4G3G Product | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16, ... | | Authors: | Hemsworth, G.R, Tamura, K, Dejean, G, Rogers, T.E, Pudlo, N.A, Urs, K, Jain, N, Martens, E.C, Brumer, H, Davies, G.J. | | Deposit date: | 2017-03-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism by which Prominent Human Gut Bacteroidetes Utilize Mixed-Linkage Beta-Glucans, Major Health-Promoting Cereal Polysaccharides.
Cell Rep, 21, 2017
|
|
6VHO
 
 | |
7KR6
 
 | |
1EW0
 
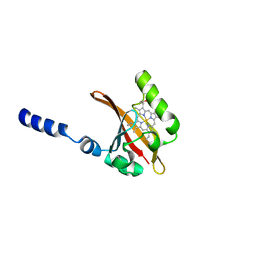 | | CRYSTAL STRUCTURE ANALYSIS OF THE SENSOR DOMAIN OF RMFIXL(FERROUS FORM) | | Descriptor: | FIXL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
1X3U
 
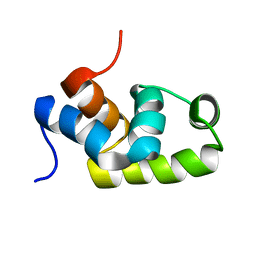 | | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium melilot | | Descriptor: | Transcriptional regulatory protein fixJ | | Authors: | Kurashima-Ito, K, Kasai, Y, Hosono, K, Tamura, K, Oue, S, Isogai, M, Ito, Y, Nakamura, H, Shiro, Y. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter
Biochemistry, 44, 2005
|
|
1D06
 
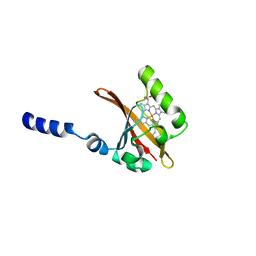 | | STRUCTURAL BASIS OF DIMERIZATION AND SENSORY MECHANISMS OF OXYGEN-SENSING DOMAIN OF RHIZOBIUM MELILOTI FIXL DETERMINED AT 1.4A RESOLUTION | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, nitrogen fixation regulatory protein fixL | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 1999-09-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
5H34
 
 | | Crystal structure of the C-terminal domain of methionyl-tRNA synthetase (MetRS-C) in Nanoarchaeum equitans | | Descriptor: | Methionine-tRNA ligase | | Authors: | Suzuki, H, Kaneko, A, Yamamoto, T, Nambo, M, Umehara, T, Yoshida, H, Park, S.Y, Tamura, K. | | Deposit date: | 2016-10-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Binding Properties of Split tRNA to the C-terminal Domain of Methionyl-tRNA Synthetase of Nanoarchaeum equitans.
J. Mol. Evol., 84, 2017
|
|
7FG6
 
 | |
5Z5E
 
 | |
1PE6
 
 | | REFINED X-RAY STRUCTURE OF PAPAIN(DOT)E-64-C COMPLEX AT 2.1-ANGSTROMS RESOLUTION | | Descriptor: | METHANOL, N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-2-METHYL-BUTANE, PAPAIN | | Authors: | Yamamoto, D, Matsumoto, K, Ohishi, H, Ishida, T, Inoue, M, Kitamura, K, Mizuno, H. | | Deposit date: | 1991-05-14 | | Release date: | 1993-04-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined x-ray structure of papain.E-64-c complex at 2.1-A resolution.
J.Biol.Chem., 266, 1991
|
|
