3STI
 
 | | Crystal structure of the protease domain of DegQ from Escherichia coli | | Descriptor: | Protease degQ | | Authors: | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
3STJ
 
 | | Crystal structure of the protease + PDZ1 domain of DegQ from Escherichia coli | | Descriptor: | Protease degQ, peptide (UNK) | | Authors: | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
4A8D
 
 | | DegP dodecamer with bound OMP | | Descriptor: | OUTER MEMBRANE PROTEIN C, PERIPLASMIC SERINE ENDOPROTEASE DEGP | | Authors: | Malet, H, Krojer, T, Sawa, J, Schafer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
3CS0
 
 | | Crystal structure of DegP24 | | Descriptor: | Periplasmic serine endoprotease DegP, pentapeptide | | Authors: | Krojer, T, Sawa, J, Schaefer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | Deposit date: | 2008-04-08 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
4A8B
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozymes | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8C
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with a binding peptide | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A8A
 
 | | Asymmetric cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozyme | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.2 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A9G
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
3MH5
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | DIISOPROPYL PHOSPHONATE, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MH6
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | DIISOPROPYL PHOSPHONATE, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MH4
 
 | |
3MH7
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | 5-mer peptide, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.961 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
1Z44
 
 | | Crystal structure of oxidized YqjM from Bacillus subtilis complexed with p-nitrophenol | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-NITROPHENOL, Probable NADH-dependent flavin oxidoreductase yqjM, ... | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z48
 
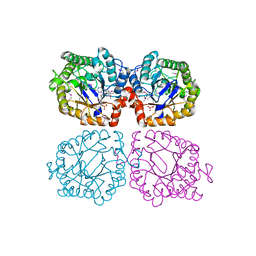 | | Crystal structure of reduced YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z41
 
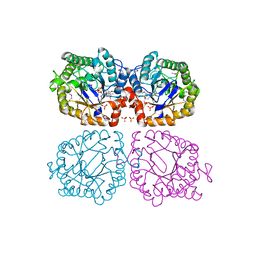 | | Crystal structure of oxidized YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM, SULFATE ION | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z42
 
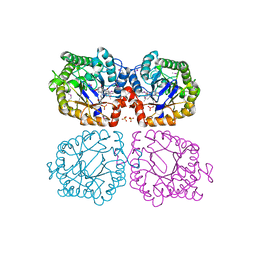 | | Crystal structure of oxidized YqjM from Bacillus subtilis complexed with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, Probable NADH-dependent flavin oxidoreductase yqjM, ... | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
2ZFU
 
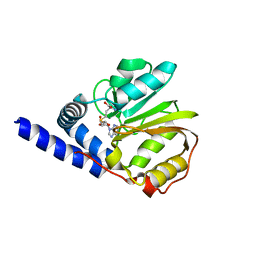 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
3W0I
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)pentan-3-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0G
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[2-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)propan-2-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0H
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[4-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)heptan-4-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
3W0J
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[2-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)propan-2-yl]-2-methylphenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
2R3U
 
 | | Crystal structure of the PDZ deletion mutant of DegS | | Descriptor: | Protease degS | | Authors: | Clausen, T, Kurzbauer, R. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the sigmaE stress response by DegS: how the PDZ domain keeps the protease inactive in the resting state and allows integration of different OMP-derived stress signals upon folding stress.
Genes Dev., 21, 2007
|
|
2R3Y
 
 | |
2ZLE
 
 | | Cryo-EM structure of DegP12/OMP | | Descriptor: | Outer membrane protein C, Protease do | | Authors: | Schaefer, E, Saibil, H.R. | | Deposit date: | 2008-04-09 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
3OU0
 
 | | re-refined 3CS0 | | Descriptor: | Periplasmic serine endoprotease DegP, heptapeptide, pentapeptide | | Authors: | Sauer, R.T, Grant, R.A, Kim, S. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Cell(Cambridge,Mass.), 145, 2011
|
|
