2I9K
 
 | | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI | | Descriptor: | 5'-D(*T*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3', Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Youngblood, B, Shieh, F.K, De Los Rios, S, Perona, J.J, Reich, N.O. | | Deposit date: | 2006-09-05 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Extrahelical Base Destabilization Enhances Sequence Discrimination of DNA Methyltransferase M.HhaI
J.Mol.Biol., 362, 2006
|
|
2ZCJ
 
 | | Ternary structure of the Glu119Gln M.HhaI, C5-Cytosine DNA methyltransferase, with unmodified DNA and AdoHcy | | Descriptor: | DNA (5'-D(*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), Modification methylase HhaI, ... | | Authors: | Shieh, F.K, Reich, N.O. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | AdoMet-dependent Methyl-transfer: Glu(119) Is Essential for DNA C5-Cytosine Methyltransferase M.HhaI
J.Mol.Biol., 373, 2007
|
|
2B0E
 
 | | EcoRV Restriction Endonuclease/GAAUTC/Ca2+ | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*AP*(DU)P*TP*CP*TP*T)-3', CALCIUM ION, Type II restriction enzyme EcoRV | | Authors: | Hiller, D.A, Rodriguez, A.M, Perona, J.J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-cognate Enzyme-DNA Complex: Structural and Kinetic Analysis of EcoRV Endonuclease Bound to the EcoRI Recognition Site GAATTC
J.Mol.Biol., 354, 2005
|
|
2B0D
 
 | | EcoRV Restriction Endonuclease/GAATTC/Ca2+ | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*AP*TP*TP*CP*TP*T)-3', CALCIUM ION, Type II restriction enzyme EcoRV | | Authors: | Hiller, D.A, Rodriguez, A.M, Perona, J.J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-cognate Enzyme-DNA Complex: Structural and Kinetic Analysis of EcoRV Endonuclease Bound to the EcoRI Recognition Site GAATTC
J.Mol.Biol., 354, 2005
|
|
6PBD
 
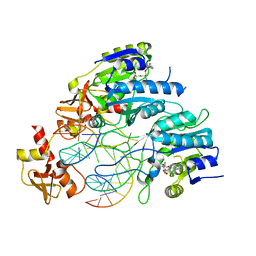 | | DNA N6-Adenine Methyltransferase CcrM In Complex with Double-Stranded DNA Oligonucleotide Containing Its Recognition Sequence GAATC | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*AP*TP*TP*CP*AP*AP*TP*GP*AP*AP*TP*CP*CP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*GP*GP*AP*TP*TP*CP*AP*TP*TP*GP*AP*AP*TP*C)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Woodcock, C.B. | | Deposit date: | 2019-06-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.343 Å) | | Cite: | The cell cycle-regulated DNA adenine methyltransferase CcrM opens a bubble at its DNA recognition site.
Nat Commun, 10, 2019
|
|
1BUA
 
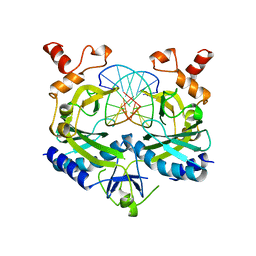 | |
1BSU
 
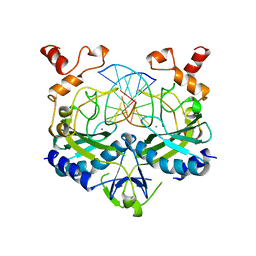 | |
2HR1
 
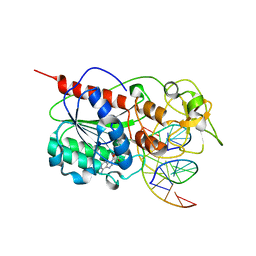 | |
2Z6A
 
 | |
2Z6Q
 
 | |
2Z6U
 
 | |
