7T1Q
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid | | Descriptor: | ACETATE ION, SUCCINIC ACID, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-02 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid.
To be Published
|
|
8F8O
 
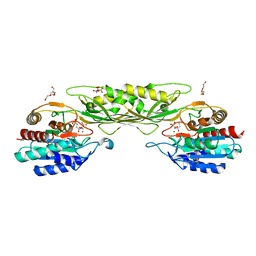 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CITRIC ACID, SUCCINIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids
To Be Published
|
|
5I4C
 
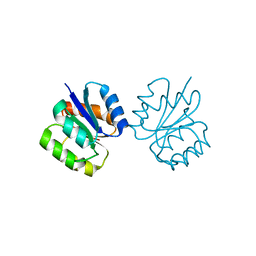 | | Crystal structure of non-phosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli | | Descriptor: | Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Ruan, J, Pshenychnyi, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli.
Protein Sci., 25, 2016
|
|
7RJ3
 
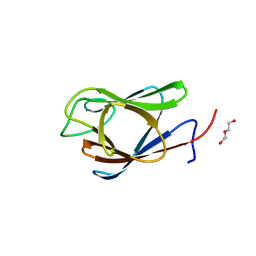 | | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycogen accumulation regulator GarA | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the Forkhead Associated (FHA) Domain of the Glycogen Accumulation Regulator (GarA) from Mycobacterium tuberculosis.
To Be Published
|
|
7KOS
 
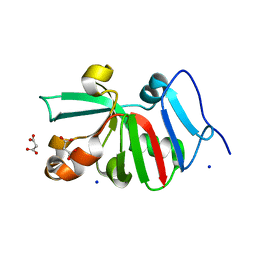 | | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | FORMIC ACID, MALONIC ACID, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7KP2
 
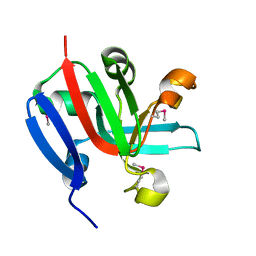 | | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin | | Descriptor: | L-NEOPTERIN, Putative Pterin Binding Protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Pshenychnyi, S, Dubrovska, I, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | High Resolution Crystal Structure of Putative Pterin Binding Protein (PruR) from Vibrio cholerae O1 biovar El Tor str. N16961 in Complex with Neopterin.
To Be Published
|
|
7KOU
 
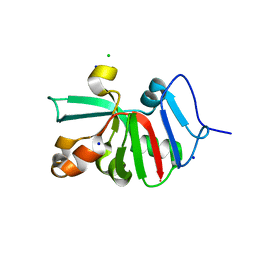 | | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
6MUK
 
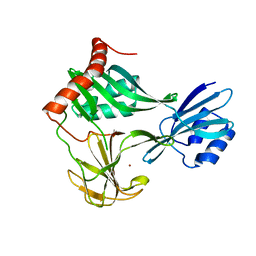 | | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae. | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae.
To Be Published
|
|
6NAU
 
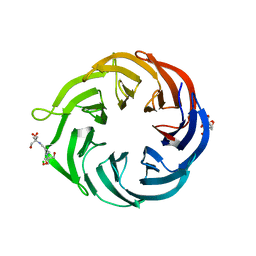 | | 1.55 Angstrom Resolution Crystal Structure of 6-phosphogluconolactonase from Klebsiella pneumoniae | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-phosphogluconolactonase, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DT3
 
 | | 1.2 Angstrom Resolution Crystal Structure of Nucleoside Triphosphatase NudI from Klebsiella pneumoniae in Complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoside triphosphatase NudI | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6E85
 
 | | 1.25 Angstrom Resolution Crystal Structure of 4-hydroxythreonine-4-phosphate Dehydrogenase from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, D-threonate 4-phosphate dehydrogenase, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6X1L
 
 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | WbbZ protein | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6WN5
 
 | | 1.52 Angstrom Resolution Crystal Structure of Transcriptional Regulator HdfR from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Transcriptional regulator HdfR | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6WN8
 
 | | 2.70 Angstrom Resolution Crystal Structure of Uracil Phosphoribosyl Transferase from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, SULFATE ION, Uracil phosphoribosyltransferase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7TL5
 
 | | Crystal structure of putative hydrolase yjcS from Klebsiella pneumoniae. | | Descriptor: | 1,2-ETHANEDIOL, Lactamase_B domain-containing protein | | Authors: | Chang, C, Endres, M, Wu, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7TZP
 
 | | Crystal Structure of Putataive Short-Chain Dehydrogenase/Reductase (FabG) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 in Complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-ACP reductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7RJJ
 
 | | Crystal Structure of the Peptidoglycan Binding Domain of the Outer Membrane Protein (OmpA) from Klebsiella pneumoniae with bound D-alanine | | Descriptor: | CHLORIDE ION, D-ALANINE, OmpA family protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DUX
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD. | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 6-phospho-alpha-glucosidase, ACETATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-22 | | Release date: | 2018-07-04 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6NDI
 
 | | Crystal Structure of the Sugar Binding Domain of LacI Family Protein from Klebsiella pneumoniae | | Descriptor: | Transcriptional regulator | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DVV
 
 | | 2.25 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Klebsiella pneumoniae in Complex with NAD and Mn2+. | | Descriptor: | 6-phospho-alpha-glucosidase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6NBG
 
 | | 2.05 Angstrom Resolution Crystal Structure of Hypothetical Protein KP1_5497 from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Glucosamine-6-phosphate deaminase, PHOSPHATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6DXN
 
 | | 1.95 Angstrom Resolution Crystal Structure of DsbA Disulfide Interchange Protein from Klebsiella pneumoniae. | | Descriptor: | TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
5VXN
 
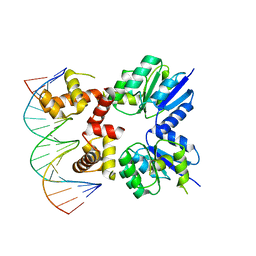 | | Structure of two RcsB dimers bound to two parallel DNAs. | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*A)-3'), Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Minasov, G, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.375 Å) | | Cite: | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
5W43
 
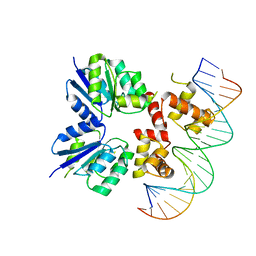 | | Structure of the two-component response regulator RcsB-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*TP*CP*CP*TP*AP*AP*AP*TP*C)-3'), Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Warwzak, Z, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
