5ZGH
 
 | | Cryo-EM structure of the red algal PSI-LHCR | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X. | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZGB
 
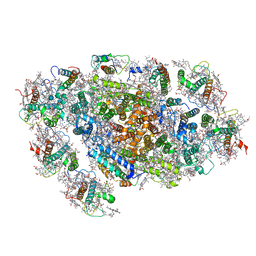 | | Cryo-EM structure of the red algal PSI-LHCR | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X. | | Deposit date: | 2018-03-08 | | Release date: | 2018-04-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
7CZL
 
 | | Structural insights into a dimeric Psb27-photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X, Huang, G, Xiao, Y. | | Deposit date: | 2020-09-09 | | Release date: | 2021-01-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insights into a dimeric Psb27-photosystem II complex from a cyanobacterium Thermosynechococcus vulcanus .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8E4C
 
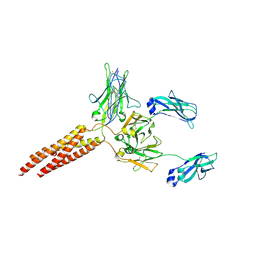 | | IgM BCR fab truncated form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain,Yellow fluorescent protein, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Dong, Y, Pi, X, Wu, H, Reth, M. | | Deposit date: | 2022-08-18 | | Release date: | 2022-11-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural principles of B cell antigen receptor assembly.
Nature, 612, 2022
|
|
6L35
 
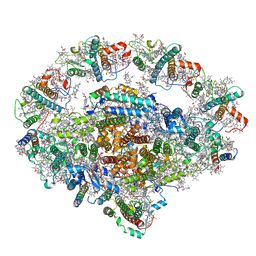 | | PSI-LHCI Supercomplex from Physcometrella patens | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, L, Yan, Q.J, Qin, X.C. | | Deposit date: | 2019-10-09 | | Release date: | 2021-02-10 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Antenna arrangement and energy-transfer pathways of PSI-LHCI from the moss Physcomitrella patens.
Cell Discov, 7, 2021
|
|
6LY5
 
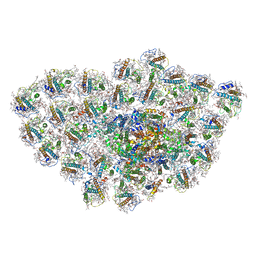 | | Organization and energy transfer in a huge diatom PSI-FCPI supercomplex | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Xiong, P, Caizhe, X. | | Deposit date: | 2020-02-13 | | Release date: | 2020-10-21 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structural basis for energy transfer in a huge diatom PSI-FCPI supercomplex.
Nat Commun, 11, 2020
|
|
6IGZ
 
 | | Structure of PSI-LHCI | | Descriptor: | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Xiong, P, Xiaochun, Q. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits.
Nat Plants, 5, 2019
|
|
9CJK
 
 | | Human TMED9 octamer structure | | Descriptor: | Transmembrane emp24 domain-containing protein 9, [(2R)-2-[(E)-octadec-9-enoyl]oxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] (E)-octadec-9-enoate | | Authors: | Le, X, Xiong, P. | | Deposit date: | 2024-07-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of TMED9 oligomerization and entrapment of misfolded protein cargo in the early secretory pathway.
Sci Adv, 10, 2024
|
|
9CJL
 
 | | Molecular basis of TMED9 dodecamer | | Descriptor: | Transmembrane emp24 domain-containing protein 9, [(2R)-2-[(E)-octadec-9-enoyl]oxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] (E)-octadec-9-enoate | | Authors: | Le, X, Xiong, P. | | Deposit date: | 2024-07-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular basis of TMED9 oligomerization and entrapment of misfolded protein cargo in the early secretory pathway.
Sci Adv, 10, 2024
|
|
8DDD
 
 | |
8DDC
 
 | |
8EMA
 
 | | mouse full length B cell receptor | | Descriptor: | Anti-human Langerin 2G3 lambda chain, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Ying, D, Xiong, P, Michael, R. | | Deposit date: | 2022-09-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural principles of B cell antigen receptor assembly.
Nature, 612, 2022
|
|
7TDZ
 
 | |
4PA8
 
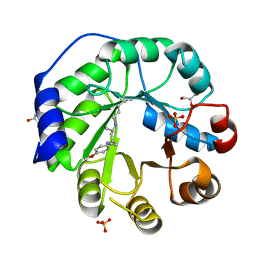 | | Crystal structure of a de novo retro-aldolase catalyzing asymmetric Michael additions, with a covalently bound product analog | | Descriptor: | (3R)-3-(4-methoxyphenyl)-5-oxohexanenitrile, GLYCEROL, SULFATE ION, ... | | Authors: | Beck, T, Garrabou Pi, X, Hilvert, D. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Promiscuous De Novo Retro-Aldolase Catalyzes Asymmetric Michael Additions via Schiff Base Intermediates.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7WOT
 
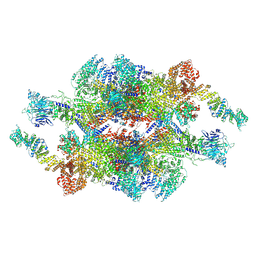 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOO
 
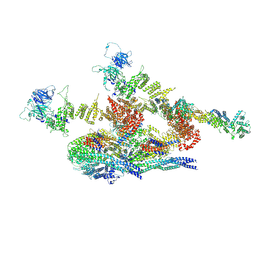 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WO9
 
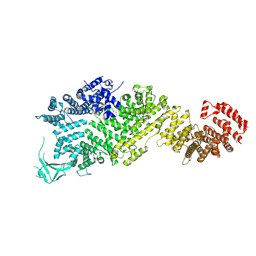 | | Cryo-EM structure of full-length Nup188 | | Descriptor: | Nucleoporin NUP188 | | Authors: | Zhao, L, Li, Z.Q, Sui, S.F. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
