7K32
 
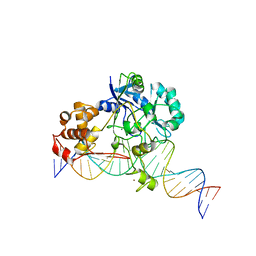 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K31
 
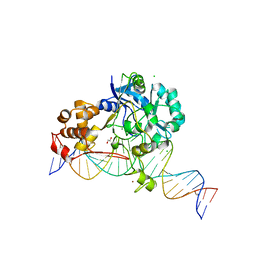 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dI at the active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (27-MER), ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K33
 
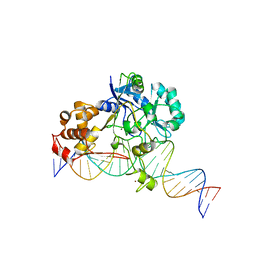 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K30
 
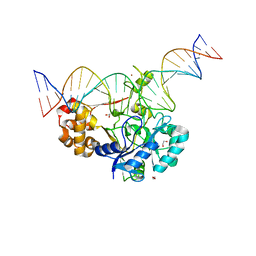 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | Descriptor: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5WFY
 
 | |
6E8C
 
 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | Authors: | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | Deposit date: | 2018-07-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
6DT1
 
 | | Crystal structure of the ligase from bacteriophage T4 complexed with DNA intermediate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction.
Nucleic Acids Res., 46, 2018
|
|
6DRT
 
 | |
6NFL
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G complexed with 2-HP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-diazinan-2-one, CHLORIDE ION, ... | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
6VOY
 
 | | Cryo-EM structure of HTLV-1 instasome | | Descriptor: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | Authors: | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | Deposit date: | 2020-02-01 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
6NFK
 
 | | Crystal Structure of the Cancer Genomic DNA Mutator APOBEC3B with loop 7 from APOBEC3G bound to iodide | | Descriptor: | 1,2-ETHANEDIOL, DNA dC->dU-editing enzyme APOBEC-3B, IODIDE ION | | Authors: | Shi, K, Orellana, K, Aihara, H. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active site plasticity and possible modes of chemical inhibition of the human DNA deaminase APOBEC3B
Faseb Bioadv, 2, 2020
|
|
7KM6
 
 | | APOBEC3B antibody 5G7 Fv-clasp | | Descriptor: | 1,2-ETHANEDIOL, 5G7 human monoclonal FAB heavy chain, 5G7 human monoclonal FAB light chain, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2020-11-02 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Characterization of a Minimal Antibody against Human APOBEC3B.
Viruses, 13, 2021
|
|
