1QFO
 
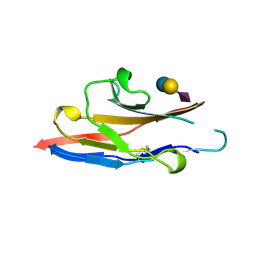 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) IN COMPLEX WITH 3'SIALYLLACTOSE | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Vinson, M, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
1QFP
 
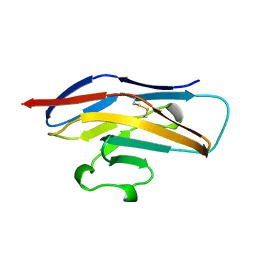 | | N-TERMINAL DOMAIN OF SIALOADHESIN (MOUSE) | | Descriptor: | PROTEIN (SIALOADHESIN) | | Authors: | May, A.P, Robinson, R.C, Burtnick, L, Crocker, P.R, Jones, E.Y. | | Deposit date: | 1999-04-12 | | Release date: | 1999-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the N-terminal domain of sialoadhesin in complex with 3' sialyllactose at 1.85 A resolution.
Mol.Cell, 1, 1998
|
|
1QDN
 
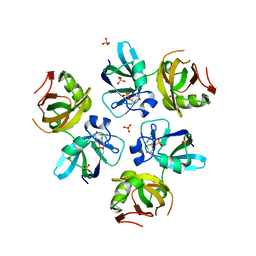 | | AMINO TERMINAL DOMAIN OF THE N-ETHYLMALEIMIDE SENSITIVE FUSION PROTEIN (NSF) | | Descriptor: | BETA-MERCAPTOETHANOL, PROTEIN (N-ETHYLMALEIMIDE SENSITIVE FUSION PROTEIN (NSF)), SULFATE ION | | Authors: | May, A.P, Misura, K.M.S, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1999-05-21 | | Release date: | 1999-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein.
Nat.Cell Biol., 1, 1999
|
|
3NUH
 
 | |
7OX8
 
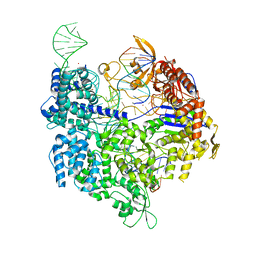 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
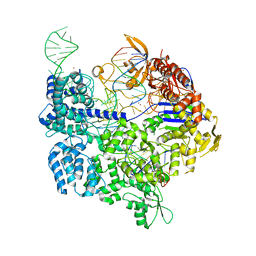 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
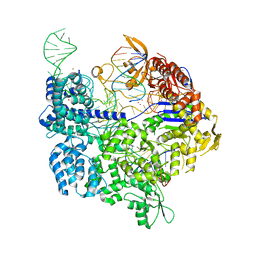 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX9
 
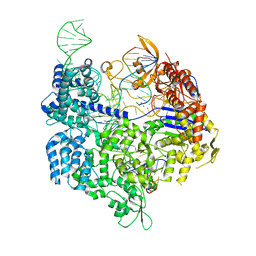 | | Target-bound SpCas9 complex with AAVS1 all-RNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Pacesa, M, Donohoue, P, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
2BVE
 
 | | Structure of the N-terminal of Sialoadhesin in complex with 2-Phenyl- Prop5Ac | | Descriptor: | SIALOADHESIN, benzyl 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosidonic acid | | Authors: | Zaccai, N.R, May, A.P, Robinson, R.C, Burtnick, L.D, Crocker, P, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2005-06-27 | | Release date: | 2006-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and in Silico Analysis of the Sialoside-Binding Characteristics of the Siglec Sialoadhesin.
J.Mol.Biol., 365, 2007
|
|
4DJB
 
 | | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors | | Descriptor: | E4-ORF3 | | Authors: | Ou, H.D, Kwiatkowski, W, Deerinck, T.J, Noske, A, Blain, K.Y, Land, H.S, Soria, C, Powers, C.J, May, A.P, Shu, X, Tsien, R.Y, Fitzpatrick, J.A.J, Long, J.A, Ellisman, M.H, Choe, S, O'Shea, C.C. | | Deposit date: | 2012-02-01 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors.
Cell(Cambridge,Mass.), 151, 2012
|
|
2XHY
 
 | | Crystal Structure of E.coli BglA | | Descriptor: | 6-PHOSPHO-BETA-GLUCOSIDASE BGLA, BROMIDE ION, SULFATE ION | | Authors: | Totir, M, Zubieta, C, Echols, N, May, A.P, Gee, C.L, nanao, M, alber, T. | | Deposit date: | 2010-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
1U9J
 
 | |
1JTH
 
 | | Crystal structure and biophysical properties of a complex between the N-terminal region of SNAP25 and the SNARE region of syntaxin 1a | | Descriptor: | SNAP25, syntaxin 1a | | Authors: | Misura, K.M.S, Gonzalez Jr, L.C, May, A.P, Scheller, R.H, Weis, W.I. | | Deposit date: | 2001-08-21 | | Release date: | 2001-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biophysical properties of a complex between the N-terminal SNARE region of SNAP25 and syntaxin 1a.
J.Biol.Chem., 276, 2001
|
|
1Z73
 
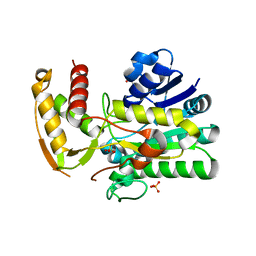 | | Crystal Structure of E. coli ArnA dehydrogenase (decarboxylase) domain, S433A mutant | | Descriptor: | GLYCEROL, SULFATE ION, protein ArnA | | Authors: | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | Deposit date: | 2005-03-24 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
1Z7E
 
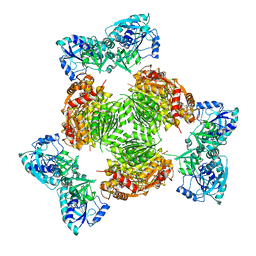 | | Crystal structure of full length ArnA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID, protein ArnA | | Authors: | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | Deposit date: | 2005-03-24 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
1Z75
 
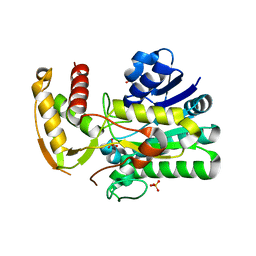 | | Crystal Structure of ArnA dehydrogenase (decarboxylase) domain, R619M mutant | | Descriptor: | GLYCEROL, SULFATE ION, protein ArnA | | Authors: | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | Deposit date: | 2005-03-24 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
1Z74
 
 | |
1YRW
 
 | |
1Z7B
 
 | |
6U20
 
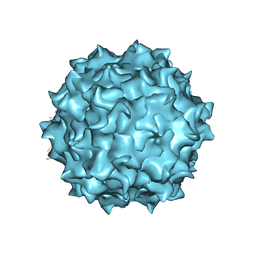 | | AAV8 human HEK293-produced, empty capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
Mol Ther Methods Clin Dev, 18, 2020
|
|
6U2V
 
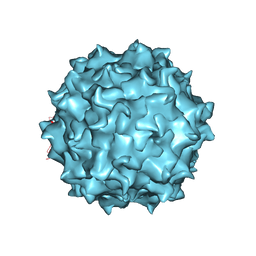 | | AAV8 Baculovirus-Sf9 produced, full capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
Mol Ther Methods Clin Dev, 18, 2020
|
|
6UBM
 
 | | AAV8 Baculovirus-Sf9 produced, empty capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-09-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
To Be Published, 18, 2020
|
|
6PWA
 
 | | AAV8 human HEK293-produced, full capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
Mol Ther Methods Clin Dev, 18, 2020
|
|
3N6Q
 
 | | Crystal structure of YghZ from E. coli | | Descriptor: | MAGNESIUM ION, YghZ aldo-keto reductase | | Authors: | Zubieta, C, Totir, M, Echols, N, May, A, Alber, T. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
3NBU
 
 | | Crystal structure of pGI glucosephosphate isomerase | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase | | Authors: | Alber, T, Zubieta, C, Totir, M, May, A, Echols, N. | | Deposit date: | 2010-06-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
