1HD9
 
 | | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif | | Descriptor: | BOWMAN-BIRK INHIBITOR DERIVED PEPTIDE | | Authors: | Brauer, A.B.E, Kelly, G, McBride, J.D, Cooke, R.M, Matthews, S.J, Leatherbarrow, R.J. | | Deposit date: | 2000-11-13 | | Release date: | 2001-03-29 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Bowman-Birk Inhibitor Reactive Site Loop Sequence Represents an Independent Structural Beta-Hairpin Motif
J.Mol.Biol., 306, 2001
|
|
8C4A
 
 | |
6FI7
 
 | | E.coli Sigma factor S (RpoS) Region 4 | | Descriptor: | RNA polymerase sigma factor RpoS | | Authors: | Liu, B, Matthews, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | T7 phage factor required for managing RpoS inEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2BEY
 
 | |
4CL1
 
 | | The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors | | Descriptor: | NON-STRUCTURAL PROTEIN 5A, SULFATE ION, ZINC ION | | Authors: | Lambert, S.M, Langley, D.R, Garnett, J.A, Angell, R, Hedgethorne, K, Meanwell, N.A, Matthews, S.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of Ns5A Domain 1 from Genotype 1A Reveals New Clues to the Mechanism of Action for Dimeric Hcv Inhibitors.
Protein Sci., 23, 2014
|
|
1GM2
 
 | |
2XSK
 
 | | E. coli curli protein CsgC - SeCys | | Descriptor: | ACETATE ION, CSGC | | Authors: | Salgado, P.S, Taylor, J.D, Cota, E, Matthews, S.J. | | Deposit date: | 2010-09-29 | | Release date: | 2010-12-29 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the Usability of the Phasing Power of Diselenide Bonds: Secys Sad Phasing of Csgc Using a Non-Auxotrophic Strain.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Y2T
 
 | |
2X43
 
 | | STRUCTURAL BASIS OF MOLECULAR RECOGNITION BY SHERP AT MEMBRANE SURFACES | | Descriptor: | SHERP | | Authors: | Moore, B, Miles, A.J, Guerra, C.G, Simpson, P, Iwata, M, Wallace, B.A, Matthews, S.J, Smith, D.F, Brown, K.A. | | Deposit date: | 2010-02-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Moelcular Recognition by the Leishmania Small Hydrophilic Endoplasmic Reticulum-Associated Protein, Sherp, at Membrane Surfaces
J.Biol.Chem., 286, 2011
|
|
5CIV
 
 | | Sibling Lethal Factor Precursor - DfsB | | Descriptor: | Sibling bacteriocin | | Authors: | Taylor, J.D, Matthews, S.J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.384 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5COF
 
 | | Crystal structure of Uncharacterised protein Q1R1X2 from Escherichia coli UTI89 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Taylor, J.D, Hare, S, Matthews, S.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5COG
 
 | | Crystal structure of Yeast IRC4 | | Descriptor: | CHLORIDE ION, IRC4, PHOSPHATE ION, ... | | Authors: | Taylor, J.D, Matthews, S.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5COM
 
 | | Crystal structure of Uncharacterized Protein Q187F5 from Clostridium difficile 630 | | Descriptor: | D(-)-TARTARIC ACID, Putative conjugative transposon protein Tn1549-like, CTn5-Orf2, ... | | Authors: | Taylor, J.D, Taylor, G, Matthews, S.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-02-03 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5CQV
 
 | | Crystal structure of uncharacterized protein Q8DWV2 from Streptococcus agalactiae | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Uncharacterized protein | | Authors: | Taylor, J.D, Hare, S, Matthews, S.J. | | Deposit date: | 2015-07-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the DfsB Protein Family Suggest a Cationic, Helical Sibling Lethal Factor Peptide.
J.Mol.Biol., 428, 2016
|
|
5DFK
 
 | |
2N59
 
 | | Solution Structure of R. palustris CsgH | | Descriptor: | Putative uncharacterized protein CsgH | | Authors: | Hawthorne, W.J, Taylor, J.D, Escalera-Maurer, A, Lambert, S, Koch, M, Scull, N, Sefer, L, Xu, Y, Matthews, S.J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-05-11 | | Method: | SOLUTION NMR | | Cite: | Electrostatically-guided inhibition of Curli amyloid nucleation by the CsgC-like family of chaperones.
Sci Rep, 6, 2016
|
|
2MP2
 
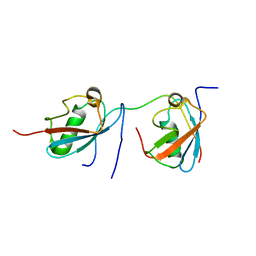 | | Solution structure of SUMO dimer in complex with SIM2-3 from RNF4 | | Descriptor: | E3 ubiquitin-protein ligase RNF4, Small ubiquitin-related modifier 3 | | Authors: | Xu, Y, Plechanovov, A, Simpson, P, Marchant, J, Leidecker, O, Sebastian, K, Hay, R.T, Matthews, S.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Nat Commun, 5, 2014
|
|
2NC9
 
 | | Apo solution structure of Hop TPR2A | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Darby, J.F, Vidler, L.R, Simpson, P.J, Matthews, S.J, Sharp, S.Y, Pearl, L.H, Hoelder, S, Workman, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hop TPR2A domain and investigation of target druggability by NMR, biochemical and in silico approaches.
Sci Rep, 10, 2020
|
|
2MA9
 
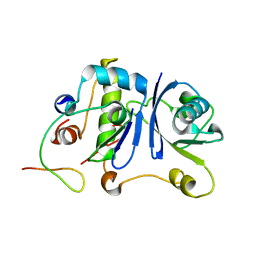 | | HIV-1 Vif SOCS-box and Elongin BC solution structure | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Lu, Z, Bergeron, J.R, Atkinson, R.A, Schaller, T, Veselkov, D.A, Oregioni, A, Yang, Y, Matthews, S.J, Malim, M.H, Sanderson, M.R. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the HIV-1 Vif SOCS-box-ElonginBC interaction.
OPEN BIOLOGY, 3, 2013
|
|
1SJQ
 
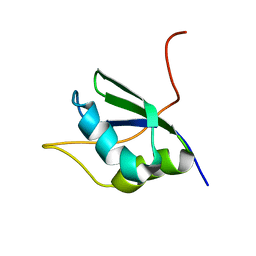 | | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1SJR
 
 | | NMR Structure of RRM2 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1E5U
 
 | | NMR Representative Structure of Intimin-190 (Int190) from Enteropathogenic E. coli | | Descriptor: | INTIMIN | | Authors: | Prasannan, S, Matthews, S.J, Batchelor, M, Daniell, S, Reece, S, Frankel, G, Dougan, G, Connerton, I, Bloomberg, G. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Recognition of the Translocated Intimin Receptor (Tir) by Intimin from Enteropathogenic E. Coli
Embo J., 19, 2000
|
|
2MPV
 
 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
8AG0
 
 | | Crystal structure of mutant PRELID3a-TRIAP1 complex - R53E | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3A, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Milara, X, Perez-Dorado, J.I, Matthews, S.J. | | Deposit date: | 2022-07-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An intermolecular hydrogen bonded network in the PRELID-TRIAP protein family plays a role in lipid sensing.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
4XZS
 
 | | Crystal Structure of TRIAP1-MBP fusion | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Abid-Ali, F, Perez-Dorado, I, Matthews, S.J. | | Deposit date: | 2015-02-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
