4MGF
 
 | |
4MF9
 
 | |
8HGU
 
 | |
8HM5
 
 | |
5FDN
 
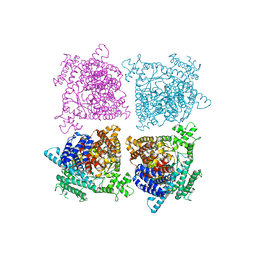 | |
8K40
 
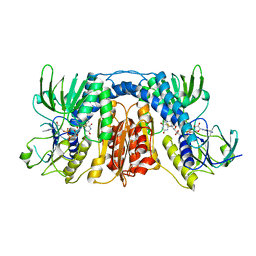 | | mercuric reductase,GbsMerA, - FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)/FAD-dependent oxidoreductase | | Authors: | Do, H. | | Deposit date: | 2023-07-17 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural basis of mercuric reductase, GbsMerA, from Gelidibacter salicanalis PAMC21136.
Sci Rep, 13, 2023
|
|
8K41
 
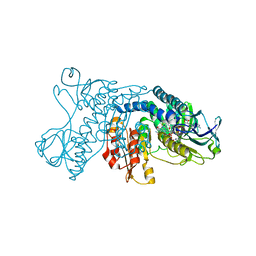 | | mercuric reductase,GbsMerA, - FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)/FAD-dependent oxidoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Do, H. | | Deposit date: | 2023-07-17 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Biochemical and structural basis of mercuric reductase, GbsMerA, from Gelidibacter salicanalis PAMC21136.
Sci Rep, 13, 2023
|
|
7XJT
 
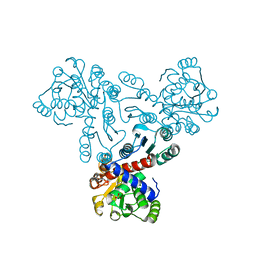 | |
7X99
 
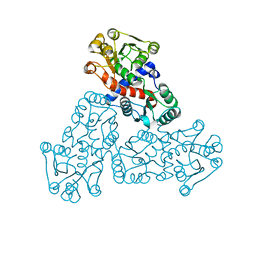 | |
7XRH
 
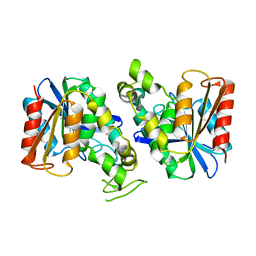 | | Feruloyl esterase from Lactobacillus acidophilus | | Descriptor: | Cinnamoyl esterase | | Authors: | Hwang, J, Lee, C.W, Lee, J.H, Do, H. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7XRI
 
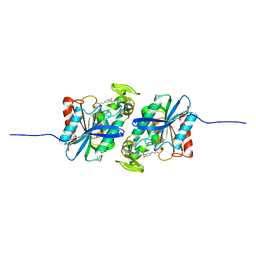 | | Feruloyl esterase mutant -S106A | | Descriptor: | Cinnamoyl esterase, ethyl (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoate | | Authors: | Hwang, J.S, Lee, J.H, Do, H, Lee, C.W. | | Deposit date: | 2022-05-10 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Feruloyl Esterase ( La Fae) from Lactobacillus acidophilus : Structural Insights and Functional Characterization for Application in Ferulic Acid Production.
Int J Mol Sci, 24, 2023
|
|
7YVT
 
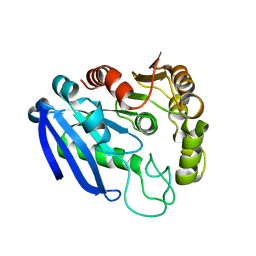 | |
7VPF
 
 | |
5C5G
 
 | |
5D6T
 
 | | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, SPHERULIN-4, ... | | Authors: | Bamford, N.C, Little, D.J, Howell, P.L. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus.
J.Biol.Chem., 290, 2015
|
|
5TSY
 
 | |
5TCB
 
 | |
7EHK
 
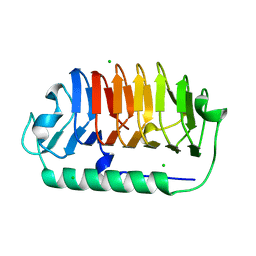 | | Crystal structure of C107S mutant of FfIBP | | Descriptor: | CHLORIDE ION, Ice-binding protein | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2021-03-29 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Importance of rigidity of ice-binding protein (FfIBP) for hyperthermal hysteresis activity and microbial survival.
Int.J.Biol.Macromol., 204, 2022
|
|
