5GOX
 
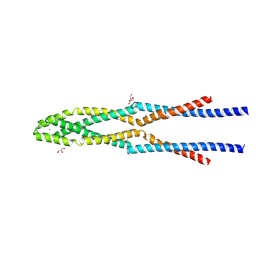 | | Eukaryotic Rad50 Functions as A Rod-shaped Dimer | | Descriptor: | DNA repair protein RAD50, GLYCEROL, ZINC ION | | Authors: | Park, Y.B, Hohl, M, Padjasek, M, Jeong, E, Jin, K.S, Krezel, A, Petrini, J.H.J, Cho, Y. | | Deposit date: | 2016-07-30 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Eukaryotic Rad50 functions as a rod-shaped dimer
Nat. Struct. Mol. Biol., 24, 2017
|
|
4P0P
 
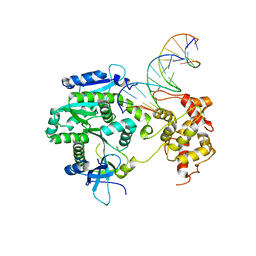 | | Crystal structure of Human Mus81-Eme1 in complex with 5'-flap DNA, and Mg2+ | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCTCAATC, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0R
 
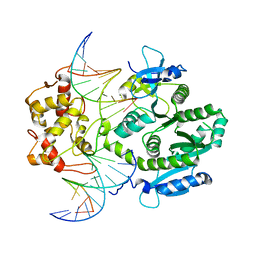 | | human Mus81-Eme1-3'flap DNA complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA ACGTGCTTACACACAGAGGTTAGGGTGAACTT, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (6.501 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0S
 
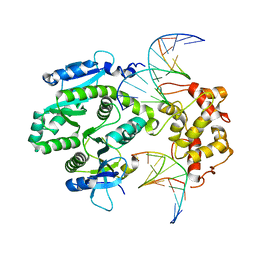 | | human Mus81-Eme1-3'flap DNA complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCT, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
6K8W
 
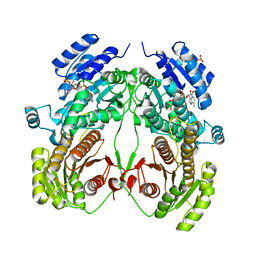 | | Crystal structure of N-domain with NADP of baterial malonyl-CoA reductase | | Descriptor: | NAD-dependent epimerase/dehydratase:Short-chain dehydrogenase/reductase SDR, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural insight into bi-functional malonyl-CoA reductase.
Environ.Microbiol., 22, 2020
|
|
6K8S
 
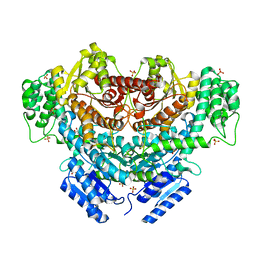 | |
6K8U
 
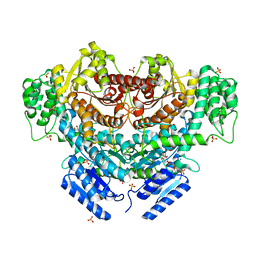 | |
6K8V
 
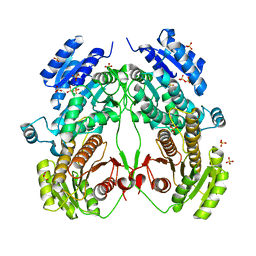 | |
4R3Z
 
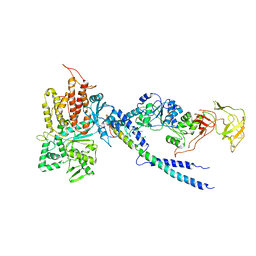 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RSJ
 
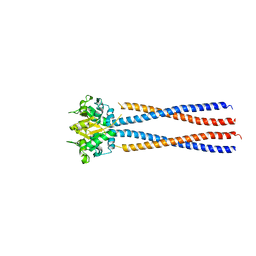 | |
4RSI
 
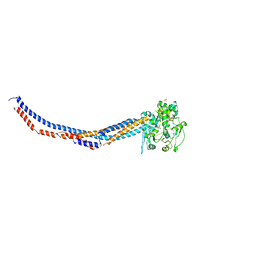 | | Yeast Smc2-Smc4 hinge domain with extended coiled coils | | Descriptor: | PHOSPHATE ION, Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Soh, Y.M, Shin, H.C, Oh, B.H. | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for SMC Rod Formation and Its Dissolution upon DNA Binding.
Mol.Cell, 57, 2015
|
|
5H5M
 
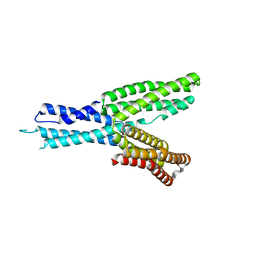 | | Crystal structure of HMP-1 M domain | | Descriptor: | Alpha-catenin-like protein hmp-1 | | Authors: | Kang, H, Bang, I, Weis, W.I, Choi, H.J. | | Deposit date: | 2016-11-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of Caenorhabditis elegans alpha-catenin reveals constitutive binding to beta-catenin and F-actin
J. Biol. Chem., 292, 2017
|
|
5ITS
 
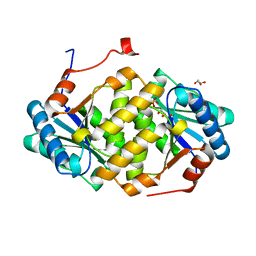 | | Crystal structure of LOG from Corynebacterium glutamicum | | Descriptor: | Cytokinin riboside 5'-monophosphate phosphoribohydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Seo, H.-G, Kim, K.-J. | | Deposit date: | 2016-03-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for cytokinin production by LOG from Corynebacterium glutamicum
Sci Rep, 6, 2016
|
|
5A1N
 
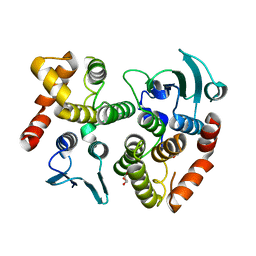 | | The crystal structure of the GST-like domains complex of EPRS-AIMP2 mutant S156D | | Descriptor: | AMINOACYL TRNA SYNTHASE COMPLEX-INTERACTING MULTIFUNCTIONAL PROTEIN 2, BIFUNCTIONAL GLUTAMATE/PROLINE--TRNA LIGASE, GLYCEROL | | Authors: | Cho, H.Y, Choi, Y.S, Kang, B.S. | | Deposit date: | 2015-05-03 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Symmetric Assembly of a Decameric Subcomplex in Human Multi-tRNA Synthetase Complex Via Interactions between Glutathione Transferase-Homology Domains and Aspartyl-tRNA Synthetase.
J.Mol.Biol., 2019
|
|
5D4Z
 
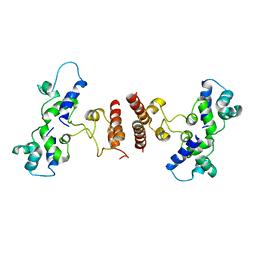 | |
5D50
 
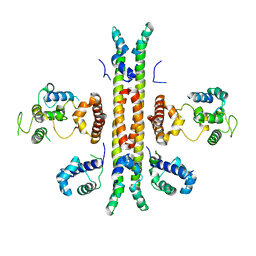 | | Crystal structure of Rep-Ant complex from Salmonella-temperate phage | | Descriptor: | Anti-repressor protein, Repressor | | Authors: | Son, S.H, Yoon, H.J, Ryu, S, Lee, H.H. | | Deposit date: | 2015-08-10 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ZQM
 
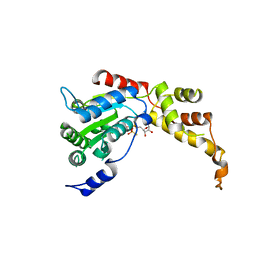 | |
6A02
 
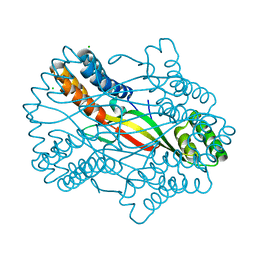 | |
6A07
 
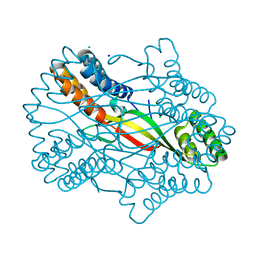 | |
6A09
 
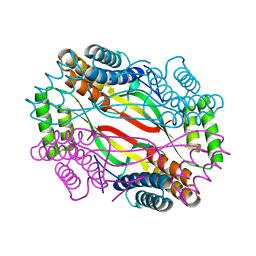 | | Salmonella Typhi YfdX in the P222 space group | | Descriptor: | YfdX protein | | Authors: | Ku, B, Lee, H.S, Kim, S.J. | | Deposit date: | 2018-06-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Physiological Exploration ofSalmonellaTyphi YfdX Uncovers Its Dual Function in Bacterial Antibiotic Stress and Virulence.
Front Microbiol, 9, 2018
|
|
5ZQL
 
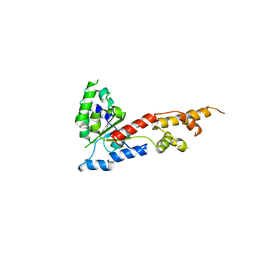 | |
4P0Q
 
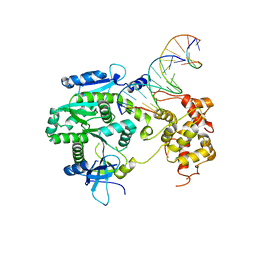 | | Crystal structure of Human Mus81-Eme1 in complex with 5'-flap DNA | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCTCAATC, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
2PYY
 
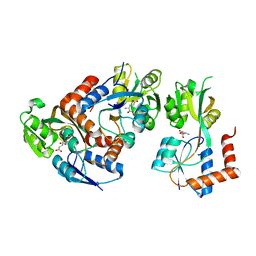 | | Crystal Structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with (L)-glutamate | | Descriptor: | GLUTAMIC ACID, Ionotropic glutamate receptor bacterial homologue | | Authors: | Lee, J.H, Kang, G.B, Lim, H.-H, Ree, M, Park, C.-S, Eom, S.H. | | Deposit date: | 2007-05-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with L-glutamate: structural dissection of the ligand interaction and subunit interface.
J.Mol.Biol., 376, 2008
|
|
2P4B
 
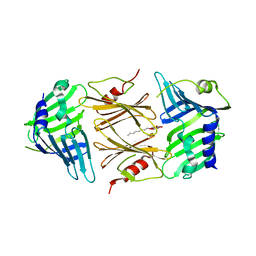 | | Crystal structure of E.coli RseB | | Descriptor: | Sigma-E factor regulatory protein rseB, octyl beta-D-glucopyranoside | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2007-03-12 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of RseB and a model of its binding mode to RseA
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6K8T
 
 | |
