8TC1
 
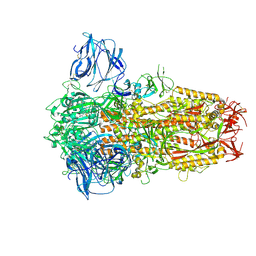 | | Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus 007 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A.R. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
8TC0
 
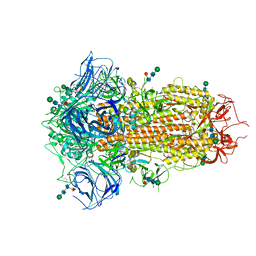 | | Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
8TC5
 
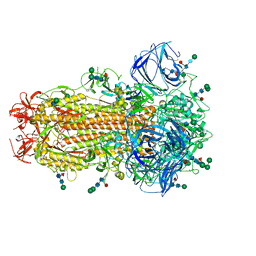 | | Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
2K7R
 
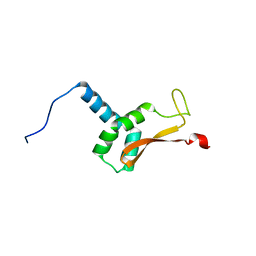 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
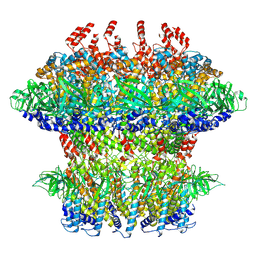
8VBX
 
 | |
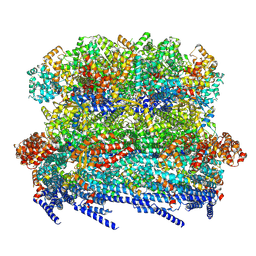
8VB4
 
 | |
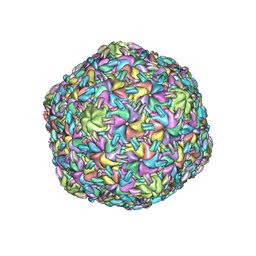
8VB2
 
 | |
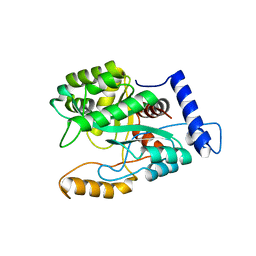
8VB0
 
 | |
4GX9
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
4GX8
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | CHLORIDE ION, DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Robinson, A, Horan, N, Xu, Z.-Q, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
