9J4G
 
 | | Crystal structure of SHMT from E. faecium with (+)-SHIN-2 | | Descriptor: | (+)-SHIN-2, Serine hydroxymethyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Hayashi, H, Murayama, K. | | Deposit date: | 2024-08-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SHIN-2 exerts potent activity against VanA-type vancomycin-resistant Enterococcus faecium in vitro by stabilizing the active site loop of serine hydroxymethyltransferase.
Arch.Biochem.Biophys., 761, 2024
|
|
1IX8
 
 | | Aspartate Aminotransferase Active Site Mutant V39F/N194A | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX7
 
 | | Aspartate Aminotransferase Active Site Mutant V39F maleate complex | | Descriptor: | Aspartate Aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
1IX6
 
 | | Aspartate Aminotransferase Active Site Mutant V39F | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
9J4H
 
 | | Crystal structure of SHMT apo form | | Descriptor: | Serine hydroxymethyltransferase, TRIETHYLENE GLYCOL | | Authors: | Murayama, K, Hayashi, H. | | Deposit date: | 2024-08-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | SHIN-2 exerts potent activity against VanA-type vancomycin-resistant Enterococcus faecium in vitro by stabilizing the active site loop of serine hydroxymethyltransferase.
Arch.Biochem.Biophys., 761, 2024
|
|
9CTX
 
 | | X-ray crystal structure of multi-drug resistant HIV-1 protease (P51) in complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | Authors: | Hayashi, H, Yedidi, R, Bulut, H, Das, D, Mitsuya, H. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Secondary amino-acid substitutions contribute to the emergence of HIV protease inhibitor resistance as directly as primary amino-acid substitutions.
To Be Published
|
|
7V3D
 
 | |
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DOY
 
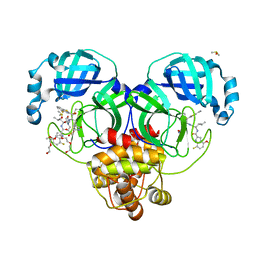 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-198 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase nsp5, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Potent and biostable inhibitors of the main protease of SARS-CoV-2.
Iscience, 25, 2022
|
|
8DOX
 
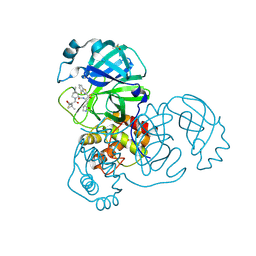 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
9ARQ
 
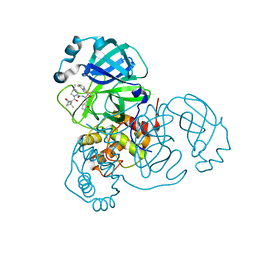 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ARS
 
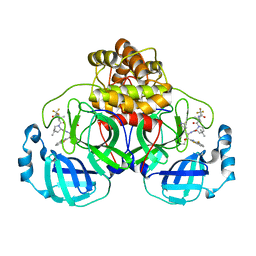 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ART
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
6OGR
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Novel HIV-1 protease inhibitors, GRL-142 and its analogs, markedly adapt to the structural plasticity of HIV-1 protease and exerts extreme potency with high genetic barrier
To Be Published
|
|
3K8U
 
 | | Crystal Structure of the Peptidase Domain of Streptococcus ComA, a Bi-functional ABC Transporter Involved in Quorum Sensing Pathway | | Descriptor: | Putative ABC transporter, ATP-binding protein ComA | | Authors: | Ishii, S, Yano, T, Ebihara, A, Okamoto, A, Manzoku, M, Hayashi, H. | | Deposit date: | 2009-10-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway
J.Biol.Chem., 285, 2010
|
|
5TYS
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
5TYR
 
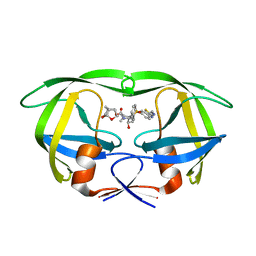 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
8UH8
 
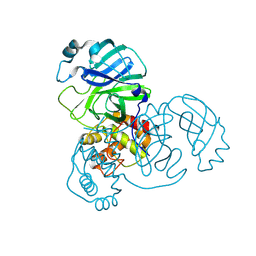 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH5
 
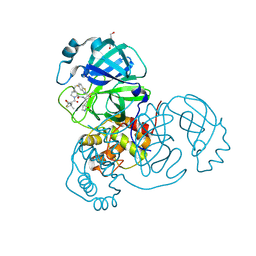 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-06 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH9
 
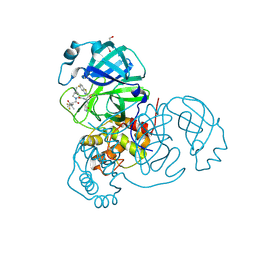 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
6UWC
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-08613 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzothiazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.
Antimicrob.Agents Chemother., 66, 2022
|
|
6UWB
 
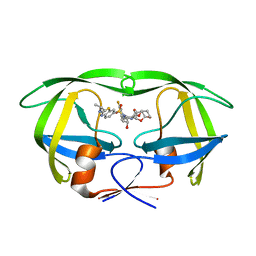 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-08513 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzothiazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.
Antimicrob.Agents Chemother., 66, 2022
|
|
5COK
 
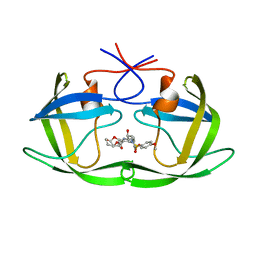 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-097 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-(4-methoxyphenyl)butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
