1QXQ
 
 | |
1QX9
 
 | |
2O4V
 
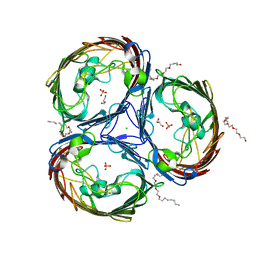 | | An arginine ladder in OprP mediates phosphate specific transfer across the outer membrane | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, T.F, Bains, M, Hancock, R.E, Strynadka, N.C. | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | An arginine ladder in OprP mediates phosphate-specific transfer across the outer membrane.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1T5M
 
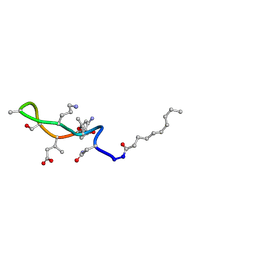 | | Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Jung, D, Rozek, A, Okon, M, Hancock, R.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin.
Chem.Biol., 11, 2004
|
|
1T5N
 
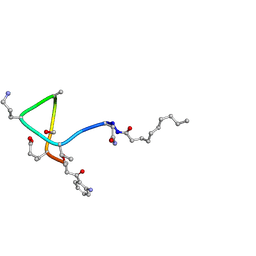 | | Structural transitions as determinants of calcium-dependent antibiotic daptomycin | | Descriptor: | DAPTOMYCIN, DECANOIC ACID | | Authors: | Jung, D, Rozek, A, Okon, M, Hancock, R.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-08-31 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin.
Chem.Biol., 11, 2004
|
|
1X7K
 
 | |
1G8C
 
 | |
1G89
 
 | |
1HR1
 
 | |
2B5K
 
 | |
1RKK
 
 | |
2LMF
 
 | | Solution structure of human LL-23 bound to membrane-mimetic micelles | | Descriptor: | Antibacterial protein LL-37 | | Authors: | Wang, G. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Dynamics, and Antimicrobial and Immune Modulatory Activities of Human LL-23 and Its Single-Residue Variants Mutated on the Basis of Homologous Primate Cathelicidins.
Biochemistry, 51, 2012
|
|
5CUL
 
 | | crystal structure of the PscU C-terminal domain | | Descriptor: | Translocation protein in type III secretion | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
5CUK
 
 | | Crystal structure of the PscP SS domain | | Descriptor: | GLYCEROL, Ruler protein | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2015-07-24 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of a Type 3 Secretion System (T3SS) Ruler Protein Suggests a Molecular Mechanism for Needle Length Sensing.
J.Biol.Chem., 291, 2016
|
|
6C41
 
 | |
5V1E
 
 | |
2N9R
 
 | | Novel antimicrobial peptide PaDBS1R1 designed from the ribosomal protein L39E from Pyrobaculum aerophilum using bioinformatics | | Descriptor: | Antimicrobial peptide PaDBS1R1 | | Authors: | Irazazabal, L.S.F, Porto, W.F, Alves, E.S.F, Matos, C.O, Hancock, R.E.W, Haney, E, Ribeiro, S.M, Liao, L.M, Ladram, A.S.F, Franco, O.L. | | Deposit date: | 2015-12-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Novel antimicrobial peptide PaDBS1R1 designed from the ribosomal protein L39E from Pyrobaculum aerophilum using bioinformatics
To be Published
|
|
