2YHF
 
 | | 1.9 Angstrom Crystal Structure of CLEC5A | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 5 MEMBER A | | Authors: | Watson, A.A, Lebedev, A.A, Murshudov, G.M, Vagin, A.A, Hall, B.A, O'Callaghan, C.A. | | Deposit date: | 2011-04-30 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Flexibility of the Macrophage Dengue Virus Receptor Clec5A: Implications for Ligand Binding and Signaling.
J.Biol.Chem., 286, 2011
|
|
1X6L
 
 | | Crystal structure of S. marcescens chitinase A mutant W167A | | Descriptor: | Chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2004-08-11 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutation of Trp167 at the -3 subsite of the chitin-binding cleft of S. marcescens chitinase A caused enhanced transglycosylation
To be Published
|
|
1X6N
 
 | | Crystal structure of S. marcescens chitinase A mutant W167A in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2004-08-11 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Muation of Trp167 at the -3 subsite of the chitin-binding cleft of S. marcescens chitinase A causes enhanced transglycosylation
To be Published
|
|
1NH6
 
 | | Structure of S. marcescens chitinase A, E315L, complex with hexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Amable, L, Madura, J.D, Pasupulati, L, Worth, C, Van Roey, P. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Family 18 chitinase-oligosaccharide substrate interaction: subsite preference and anomer selectivity of Serratia marcescens chitinase A.
Biochem.J., 376, 2003
|
|
1RD6
 
 | | Crystal Structure of S. Marcescens Chitinase A Mutant W167A | | Descriptor: | Chitinase A | | Authors: | Aronson, N.N, Halloran, B.A, Alexyev, M.F, Zhou, X.E, Wang, Y, Meehan, E.J, Chen, L. | | Deposit date: | 2003-11-05 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutation of a conserved tryptophan in the chitin-binding cleft of Serratia marcescens chitinase A enhances transglycosylation.
Biosci.Biotechnol.Biochem., 70, 2006
|
|
3ZJZ
 
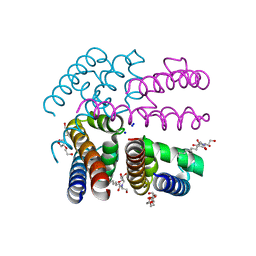 | | Open-form NavMS Sodium Channel Pore (with C-terminal Domain) | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, ION TRANSPORT PROTEIN, ... | | Authors: | Bagneris, C, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2013-01-21 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Role of the C-Terminal Domain in the Structure and Function of Tetrameric Sodium Channels.
Nat.Commun., 4, 2013
|
|
4OXS
 
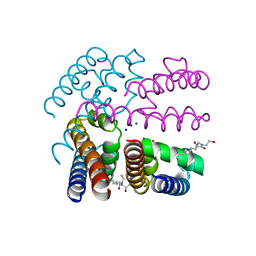 | | Structure of NavMS in complex with channel blocking compound | | Descriptor: | BROMIDE ION, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-02-06 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PA6
 
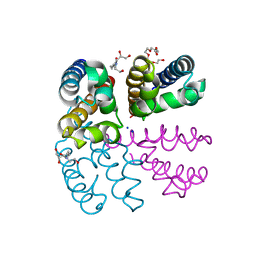 | | Structure of NavMS pore and C-terminal domain crystallised in the presence of channel blocking compound | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P30
 
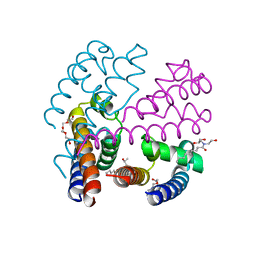 | | Structure of NavMS mutant in presence of PI1 compound | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Bagneris, C, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2014-03-05 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P2Z
 
 | | Structure of NavMS T207A/F214A | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Bagneris, C, Naylor, C.E, Wallace, B.A. | | Deposit date: | 2014-03-05 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5BZB
 
 | | NavMs voltage-gated sodium channel pore and C-terminal domain | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2015-06-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4P9O
 
 | |
4PA4
 
 | | Structure of NavMS in complex with channel blocking compound | | Descriptor: | BROMIDE ION, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PA9
 
 | | Structure of NavMS in complex with channel blocking compound | | Descriptor: | BROMIDE ION, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P9P
 
 | | Structure of NavMS in complex with channel blocking compound | | Descriptor: | BROMIDE ION, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PA3
 
 | | Structure of NavMS in complex with channel blocking compound | | Descriptor: | BROMIDE ION, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PA7
 
 | | Structure of NavMS pore and C-terminal domain crystallised in presence of channel blocking compound | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-04-07 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4X89
 
 | | NavMs voltage-gated sodium channal pore and C-terminal domain soaked with Silver nitrate | | Descriptor: | HEGA-10, Ion transport protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4X88
 
 | | E178D Selectivity filter mutant of NavMS voltage-gated pore | | Descriptor: | HEGA-10, Ion transport protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4X8A
 
 | | NavMS pore and C-terminal domain grown from protein purified in LiCl | | Descriptor: | HEGA-10, Ion transport protein, NONAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4CBC
 
 | |
3LQM
 
 | | Structure of the IL-10R2 Common Chain | | Descriptor: | GLYCEROL, Interleukin-10 receptor subunit beta, SULFATE ION | | Authors: | Yoon, S.I, Walter, M.R. | | Deposit date: | 2010-02-09 | | Release date: | 2010-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and mechanism of receptor sharing by the IL-10R2 common chain.
Structure, 18, 2010
|
|
