1L1Y
 
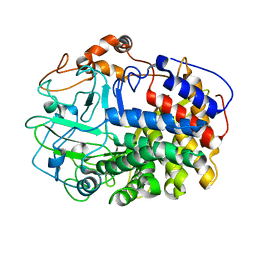 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
1L2A
 
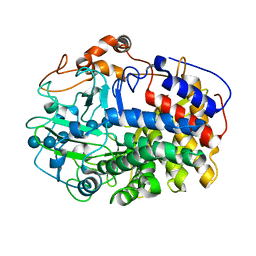 | | The Crystal Structure and Catalytic Mechanism of Cellobiohydrolase CelS, the Major Enzymatic Component of the Clostridium thermocellum cellulosome | | Descriptor: | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, cellobiohydrolase | | Authors: | Guimaraes, B.G, Souchon, H, Lytle, B.L, Wu, J.H.D, Alzari, P.M. | | Deposit date: | 2002-02-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum Cellulosome.
J.Mol.Biol., 320, 2002
|
|
3PQJ
 
 | |
2BMX
 
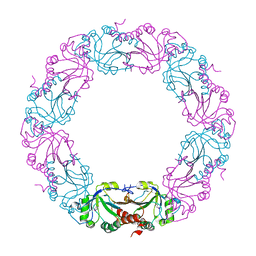 | | Mycobacterium tuberculosis AhpC | | Descriptor: | ALKYL HYDROPEROXIDASE C | | Authors: | Guimaraes, B.G, Alzari, P.M. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Mechanism of the Alkyl Hydroperoxidase Ahpc, a Key Element of the Mycobacterium Tuberculosis Defense System Against Oxidative Stress.
J.Biol.Chem., 280, 2005
|
|
4MPM
 
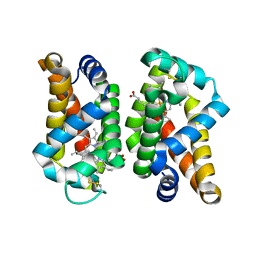 | | Wild-type human neuroglobin | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Guimaraes, B.G, Golinelli-Pimpaneau, B. | | Deposit date: | 2013-09-13 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of wild-type human brain neuroglobin reveals flexibility of the disulfide bond that regulates oxygen affinity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6OZU
 
 | |
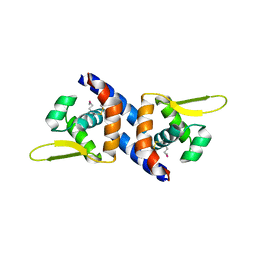
3PQK
 
 | |
6MD3
 
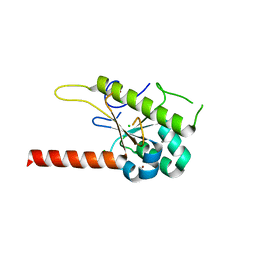 | | Structure of T. brucei RRP44 PIN domain | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Rrp44p homologue, ... | | Authors: | Guimaraes, B.G, Cesaro, G. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Trypanosoma brucei RRP44 is involved in an early stage of large ribosomal subunit RNA maturation.
RNA Biol, 16, 2019
|
|
6O7Y
 
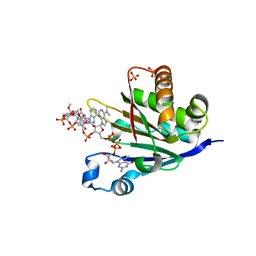 | | Trypanosoma cruzi EIF4E5 translation initiation factor in complex with cap-4 | | Descriptor: | 2-amino-9-[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(S)-({(2R,3R,4R,5R)-3-{[(R)-{[(2R,3R,4R,5R)-3-{[(S)-{[(2R,3R,4R,5R)-5-(4-amino-2-oxopyrimidin-1(2H)-yl)-3-{[(S)-hydroxy{[(2R,3R,4R,5R)-3-hydroxy-4-methoxy-5-(3-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methoxy}phosphoryl]oxy}-4-methoxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-(6-amino-9H-purin-9-yl)-4-methoxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-[6-(dimethylamino)-9H-purin-9-yl]-4-methoxytetrahydrofuran-2-yl}methoxy)(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]-7-methyl-6-oxo-6,9-dihydro-3H-purin-7-ium, Putative Eukaryotic translation initiation factor 4E type 5, SULFATE ION | | Authors: | Guimaraes, B.G, Reolon, L.W. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Trypanosoma cruzi EIF4E5 translation factor homologue in complex with mRNA cap-4.
Nucleic Acids Res., 47, 2019
|
|
6O80
 
 | |
6O7Z
 
 | | Trypanosoma cruzi EIF4E5 translation initiation factor in complex with cap-1 | | Descriptor: | 2-amino-9-[(2R,3R,4S,5R)-5-({[(R)-{[(S)-{[(R)-({(2R,3R,4R,5R)-5-[6-(dimethylamino)-9H-purin-9-yl]-3-hydroxy-4-methoxytetrahydrofuran-2-yl}methoxy)(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]-7-methyl-6-oxo-6,9-dihydro-1H-purin-7-ium, Putative Eukaryotic translation initiation factor 4E type 5, SULFATE ION | | Authors: | Guimaraes, B.G, Reolon, L.W. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Trypanosoma cruzi EIF4E5 translation factor homologue in complex with mRNA cap-4.
Nucleic Acids Res., 47, 2019
|
|
2P38
 
 | |
3BWS
 
 | | Crystal structure of the leptospiral antigen Lp49 | | Descriptor: | SULFATE ION, protein Lp49 | | Authors: | Giuseppe, P.O, Neves, F.O, Nascimento, A.L.T.O, Guimaraes, B.G. | | Deposit date: | 2008-01-10 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The leptospiral antigen Lp49 is a two-domain protein with putative protein binding function
J.Struct.Biol., 163, 2008
|
|
7TUV
 
 | |
1T13
 
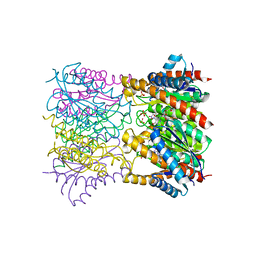 | | Crystal Structure Of Lumazine Synthase From Brucella Abortus Bound To 5-nitro-6-(D-ribitylamino)-2,4(1H,3H) pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Vega, D.R, Guimaraes, B.G, Braden, B.C, Goldbaum, F.A. | | Deposit date: | 2004-04-15 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies on Decameric Brucella spp. Lumazine Synthase: A Novel Quaternary Arrangement Evolved for a New Function?
J.Mol.Biol., 353, 2005
|
|
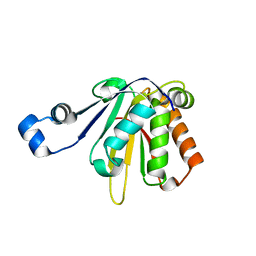
7KCJ
 
 | |
3NRK
 
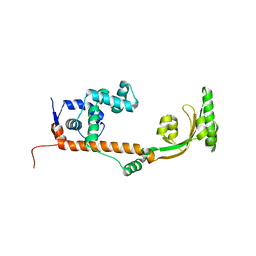 | | The crystal structure of the leptospiral hypothetical protein LIC12922 | | Descriptor: | LIC12922 | | Authors: | Giuseppe, P.O, Atzingen, M.V, Nascimento, A.L.T.O, Zanchin, N.I.T, Guimaraes, B.G. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of the leptospiral hypothetical protein LIC12922 reveals homology with the periplasmic chaperone SurA.
J.Struct.Biol., 173, 2011
|
|
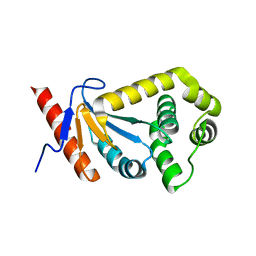
2REM
 
 | |
2PO2
 
 | |
2PNZ
 
 | |
3H79
 
 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
3ITJ
 
 | | Crystal structure of Saccharomyces cerevisiae thioredoxin reductase 1 (Trr1) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 1 | | Authors: | Oliveira, M.A, Discola, K.F, Alves, S.V, Medrano, F.J, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system.
Biochemistry, 49, 2010
|
|
1XN1
 
 | | Crystal Structure Of Lumazine Synthase From Brucella Abortus (Orthorhombic Form At 3.05 Angstroms) | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Klinke, S, Zylberman, V, Vega, D.R, Guimaraes, B.G, Braden, B.C, Goldbaum, F.A. | | Deposit date: | 2004-10-04 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystallographic studies on Decameric Brucella spp. Lumazine Synthase: A Novel Quaternary Arrangement Evolved for a New Function?
J.Mol.Biol., 353, 2005
|
|
1ZB9
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
1ZB8
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
