8B4T
 
 | |
8B5F
 
 | |
4E7R
 
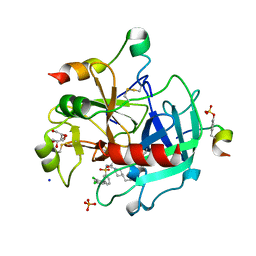 | | Thrombin in complex with 3-amidinophenylalanine inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-{[(2',4'-dichlorobiphenyl-3-yl)sulfonyl]amino}-3-oxopropyl]benzenecarboximidamide, GLYCEROL, ... | | Authors: | Ruehmann, E, Heine, A, Klebe, G. | | Deposit date: | 2012-03-19 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New 3-amidinophenylalanine-derived inhibitors of matriptase
MEDCHEMCOMM, 3, 2012
|
|
6YI7
 
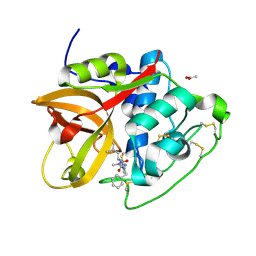 | | Structure of cathepsin B1 from Schistosoma mansoni (SmCB1) in complex with an azanitrile inhibitor | | Descriptor: | 1-[(2~{S})-1-[[iminomethyl(methyl)amino]-methyl-amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-3-(phenylmethyl)urea, ACETATE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Jilkova, A, Rezacova, P, Pachl, P, Fanfrlik, J, Rubesova, P, Guetschow, M, Mares, M. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Azanitrile Inhibitors of the SmCB1 Protease Target Are Lethal to Schistosoma mansoni : Structural and Mechanistic Insights into Chemotype Reactivity.
Acs Infect Dis., 7, 2021
|
|
6F5M
 
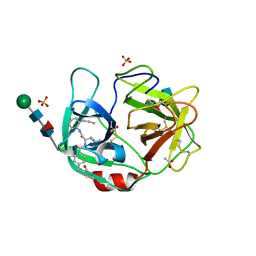 | | Crystal structure of highly glycosylated human leukocyte elastase in complex with a thiazolidinedione inhibitor | | Descriptor: | 5-[[4-[[(2~{S})-4-methyl-1-oxidanylidene-1-[(2-propylphenyl)amino]pentan-2-yl]carbamoyl]phenyl]methyl]-2-oxidanylidene-1,3-thiazol-1-ium-4-olate, ACETATE ION, Neutrophil elastase, ... | | Authors: | Hochscherf, J, Pietsch, M, Tieu, W, Kuan, K, Hautmann, S, Abell, A, Guetschow, M, Niefind, K. | | Deposit date: | 2017-12-01 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of highly glycosylated human leukocyte elastase in complex with an S2' site binding inhibitor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8RJZ
 
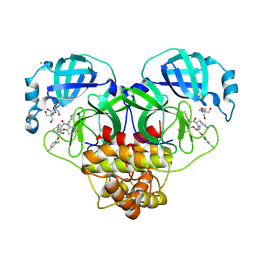 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GUE-3801 (compound 80 in publication) | | Descriptor: | (7~{S})-6-[2-[2,4-bis(chloranyl)phenoxy]ethanoyl]-14-fluoranyl-10-(iminomethyl)-9-methyl-7-(phenylmethyl)-2-oxa-6,9,10-triazabicyclo[10.4.0]hexadeca-1(12),13,15-trien-8-one, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RJY
 
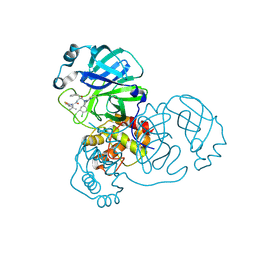 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3899 (compound 58 in publication) | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(4-chlorophenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]thiophene-2-carboxamide | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8RJV
 
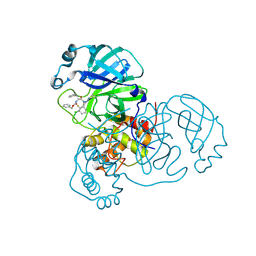 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3778 (compound 12 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(3-chloranyl-2-fluoranyl-phenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
8QDC
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3642 (compound 1 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(2~{S})-1-[[iminomethyl-(phenylmethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of an active-site titrant for SARS-CoV-2 main protease as an indispensable tool for evaluating enzyme kinetics.
Acta Pharm Sin B, 14, 2024
|
|
