4MDP
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens in complex with glucose | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3NRK
 
 | | The crystal structure of the leptospiral hypothetical protein LIC12922 | | Descriptor: | LIC12922 | | Authors: | Giuseppe, P.O, Atzingen, M.V, Nascimento, A.L.T.O, Zanchin, N.I.T, Guimaraes, B.G. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of the leptospiral hypothetical protein LIC12922 reveals homology with the periplasmic chaperone SurA.
J.Struct.Biol., 173, 2011
|
|
3BWS
 
 | | Crystal structure of the leptospiral antigen Lp49 | | Descriptor: | SULFATE ION, protein Lp49 | | Authors: | Giuseppe, P.O, Neves, F.O, Nascimento, A.L.T.O, Guimaraes, B.G. | | Deposit date: | 2008-01-10 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The leptospiral antigen Lp49 is a two-domain protein with putative protein binding function
J.Struct.Biol., 163, 2008
|
|
5JH1
 
 | | Crystal structure of the apo form of AKR4C7 from maize | | Descriptor: | Aldose reductase, AKR4C7 | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5JH2
 
 | | Crystal structure of the holo form of AKR4C7 from maize | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-2'-5'-DIPHOSPHATE, Aldose reductase, ... | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
4MDO
 
 | | Crystal structure of a GH1 beta-glucosidase from the fungus Humicola insolens | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Souza, T.A.C.B, Souza, F.H.M, Zanphorlin, L.M, Machado, C.B, Ward, R.J, Jorge, J.A, Furriel, R.P.M, Murakami, M.T. | | Deposit date: | 2013-08-23 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for glucose tolerance in GH1 beta-glucosidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4DAR
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
3RLH
 
 | | Crystal structure of a class II phospholipase D from Loxosceles intermedia venom | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Giuseppe, P.O, Ullah, A, Veiga, S.S, Murakami, M.T, Arni, R.K. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulphide bridge.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
3RLG
 
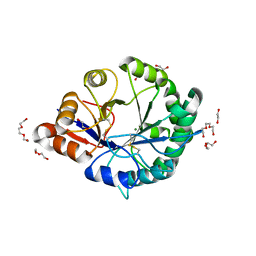 | | Crystal structure of Loxosceles intermedia phospholipase D isoform 1 H12A mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Giuseppe, P.O, Ullah, A, Veiga, S.S, Murakami, M.T, Arni, R.K. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of a class II phospholipase D from Loxosceles intermedia venom.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4KB3
 
 | |
4D9H
 
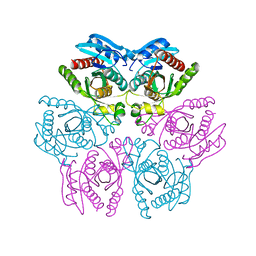 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with adenosine | | Descriptor: | ADENOSINE, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAO
 
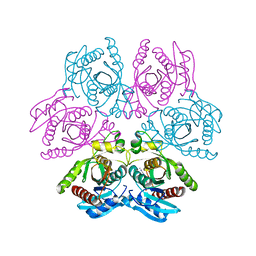 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with adenine | | Descriptor: | ADENINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAN
 
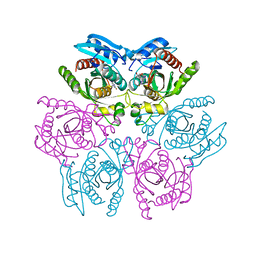 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
6D25
 
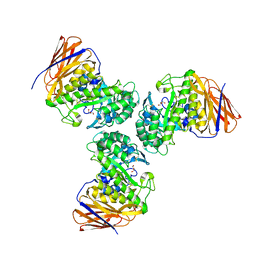 | | Crystal structure of the GH51 arabinofuranosidase from Xanthomonas axonopodis pv. citri | | Descriptor: | Alpha-L-arabinosidase, GLYCEROL | | Authors: | Santos, C.R, Morais, M.A.B, Tonoli, C.C.C, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-04-13 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The mechanism by which a distinguishing arabinofuranosidase can cope with internal di-substitutions in arabinoxylans.
Biotechnol Biofuels, 11, 2018
|
|
4DAB
 
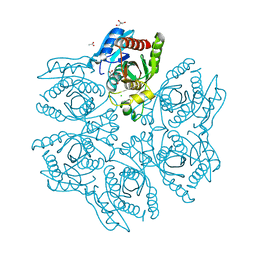 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with hypoxanthine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DA7
 
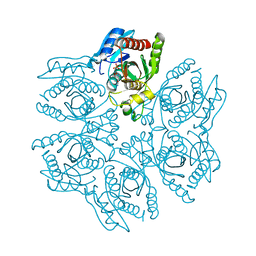 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with aciclovir | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
5JGY
 
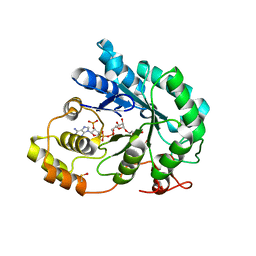 | | Crystal structure of maize AKR4C13 in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]oxy}butanoic acid (non-preferred name), Aldose reductase, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of maize AKR4C13 in P21 space group
To Be Published
|
|
5JGW
 
 | | Crystal structure of maize AKR4C13 in complex with NADP and acetate | | Descriptor: | ACETATE ION, Aldose reductase, AKR4C13, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of maize AKR4C13 in complex with NADP and acetate
To Be Published
|
|
5U4Z
 
 | | Crystal structure of citrus MAF1 in space group P 31 2 1 | | Descriptor: | Repressor of RNA polymerase III transcription, SULFATE ION | | Authors: | Soprano, A.S, Giuseppe, P.O, Nascimento, A.F.Z, Benedetti, C.E, Murakami, M.T. | | Deposit date: | 2016-12-06 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of citrus MAF1 in space group P 31 2 1
To Be Published
|
|
5U50
 
 | |
6MOY
 
 | |
6MP7
 
 | | Crystal structure of the E257A mutant of BlMan5B in complex with GlcNAc (soaking) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BlMan5B, CITRATE ANION, ... | | Authors: | Lorizolla-Cordeiro, R, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2018-10-05 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | N-glycan Utilization by Bifidobacterium Gut Symbionts Involves a Specialist beta-Mannosidase.
J. Mol. Biol., 431, 2019
|
|
6MP2
 
 | |
6MPC
 
 | |
6MPA
 
 | |
