4QYJ
 
 | |
1CKV
 
 | |
3IHM
 
 | |
4F07
 
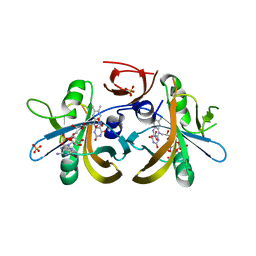 | | Structure of the Styrene Monooxygenase Flavin Reductase (SMOB) from Pseudomonas putida S12 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NITRATE ION, SULFATE ION, ... | | Authors: | Sazinsky, M.H, Morrison, E, Kantz, A, Gassner, G. | | Deposit date: | 2012-05-03 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Styrene Monooxygenase Reductase: New Insight into the FAD-Transfer Reaction.
Biochemistry, 52, 2013
|
|
7JRQ
 
 | | Crystallographically Characterized De Novo Designed Mn-Diphenylporphyrin Binding Protein | | Descriptor: | CHLORIDE ION, GLYCEROL, MPP1, ... | | Authors: | Mann, S.I, DeGrado, W.F. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | De Novo Design, Solution Characterization, and Crystallographic Structure of an Abiological Mn-Porphyrin-Binding Protein Capable of Stabilizing a Mn(V) Species.
J.Am.Chem.Soc., 143, 2021
|
|
