4YFX
 
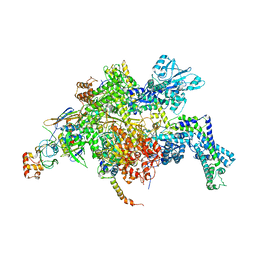 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YFK
 
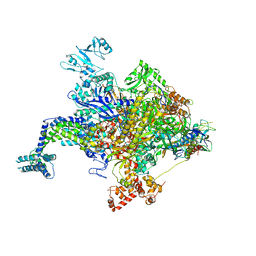 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4YFN
 
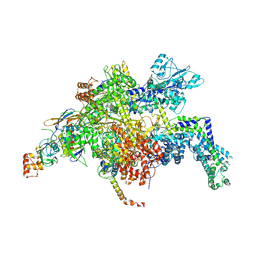 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4UCU
 
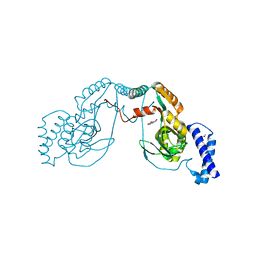 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCV
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-methoxy-2,3-dimethylquinoxalin-5-ol, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCR
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxy-2-methylquinoline-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCO
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 7-amino-2-tert-butyl-4-(1H-pyrrol-2-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCS
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5-amino-3-(furan-2-yl)-1H-1,2,4-triazole-1-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4B1F
 
 | | Design of Inhibitors of Helicobacter pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty | | Descriptor: | 5-methyl-3-(1-methyl-1H-imidazol-5-yl)-7-(2-methylpropyl)-2-(naphthalen-1-ylmethyl)-2H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, D-GLUTAMIC ACID, GLUTAMATE RACEMASE | | Authors: | Basarab, G.S, Hill, P, Eyermann, C.J, Gowravaram, M, Kack, H, Osimoni, E. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Inhibitors of Helicobacter Pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles Onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
9H1D
 
 | | Crystal structure of Angiotensin-1 converting enzyme C-domain in complex with dual ACE/NEP inhibitor AD015 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-3-phenyl-2-sulfanyl-propanoyl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Mercapto-3-phenylpropanoyl Dipeptides as Dual Angiotensin-Converting Enzyme C-Domain-Selective/Neprilysin Inhibitors.
J.Med.Chem., 68, 2025
|
|
9H1A
 
 | | Crystal structure of Angiotensin-1 converting enzyme N-domain in complex with dual ACE/NEP inhibitor AD014 | | Descriptor: | (2~{S},5~{R})-5-(3-hydroxyphenyl)-1-[2-[[(2~{S})-3-(4-hydroxyphenyl)-2-sulfanyl-propanoyl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Novel Mercapto-3-phenylpropanoyl Dipeptides as Dual Angiotensin-Converting Enzyme C-Domain-Selective/Neprilysin Inhibitors.
J.Med.Chem., 68, 2025
|
|
9H1C
 
 | | Crystal structure of Angiotensin-1 converting enzyme C-domain in complex with dual ACE/NEP inhibitor AD014 | | Descriptor: | (2~{S},5~{R})-5-(3-hydroxyphenyl)-1-[2-[[(2~{S})-3-(4-hydroxyphenyl)-2-sulfanyl-propanoyl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Mercapto-3-phenylpropanoyl Dipeptides as Dual Angiotensin-Converting Enzyme C-Domain-Selective/Neprilysin Inhibitors.
J.Med.Chem., 68, 2025
|
|
9H1B
 
 | | Crystal structure of Angiotensin-1 converting enzyme N-domain in complex with dual ACE/NEP inhibitor AD015 | | Descriptor: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-3-phenyl-2-sulfanyl-propanoyl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Novel Mercapto-3-phenylpropanoyl Dipeptides as Dual Angiotensin-Converting Enzyme C-Domain-Selective/Neprilysin Inhibitors.
J.Med.Chem., 68, 2025
|
|
9H1E
 
 | |
6W44
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 4 | | Descriptor: | 5-[methyl-[(2-propoxypyridin-3-yl)methyl]amino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W4C
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 5 | | Descriptor: | 5-[[3-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-2-oxidanyl-phenyl]methylamino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W45
 
 | | Crystal structure of HAO1 in complex with biaryl acid inhibitor - compound 3 | | Descriptor: | 2-chloranyl-4-[2-[[(6-chloranyl-1~{H}-indol-2-yl)carbonyl-methyl-amino]methyl]-5-fluoranyl-phenyl]benzoic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
9D3Y
 
 | | Crystal structure of the catalytic region of human MASP-2 with specific inhibitor Compound S2 | | Descriptor: | (2S)-N-[2-(2-amino-1H-1,3-benzimidazol-5-yl)ethyl]-1-[(2R,4S)-4-phenylpiperidine-2-carbonyl]azetidine-2-carboxamide, GLYCEROL, Mannan-binding lectin serine protease 2 B chain | | Authors: | Sawyer, T.K, Wibowo, A.S, Carter, J, Moussa, S.H. | | Deposit date: | 2024-08-12 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Small molecule inhibitors of mannan-binding lectin-associated serine Proteases-2 and-3.
Eur.J.Med.Chem., 289, 2025
|
|
9D40
 
 | | Crystal structure of the catalytic region of human MASP-2 with specific inhibitor Analog 20 | | Descriptor: | GLYCEROL, Mannan-binding lectin serine protease 2 B chain, N-{[4-(2-amino-1H-imidazol-4-yl)phenyl]methyl}-2-[4-(benzenesulfonamido)phenyl]acetamide | | Authors: | Sawyer, T.K, Wibowo, A.S, Carter, J, Moussa, S.H. | | Deposit date: | 2024-08-12 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Small molecule inhibitors of mannan-binding lectin-associated serine Proteases-2 and-3.
Eur.J.Med.Chem., 289, 2025
|
|
9D17
 
 | | Crystal structure of the catalytic region of human MASP-2 with specific inhibitor Compound S1 | | Descriptor: | 2-(2-amino-1H-1,3-benzimidazol-1-yl)-N-(5-ethylpyridin-2-yl)acetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sawyer, T.K, Wibowo, A.S, Carter, J, Moussa, S.H. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small molecule inhibitors of mannan-binding lectin-associated serine Proteases-2 and-3.
Eur.J.Med.Chem., 289, 2025
|
|
9D4D
 
 | | Crystal structure of the catalytic region of human MASP-2 with specific inhibitor Compound S3 | | Descriptor: | 2-(2-amino-1H-1,3-benzimidazol-4-yl)-N-{4-[(pyridin-4-yl)methyl]phenyl}acetamide, GLYCEROL, Mannan-binding lectin serine protease 2 B chain | | Authors: | Sawyer, T.K, Wibowo, A.S, Carter, J, Moussa, S.H. | | Deposit date: | 2024-08-12 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Small molecule inhibitors of mannan-binding lectin-associated serine Proteases-2 and-3.
Eur.J.Med.Chem., 289, 2025
|
|
4WZ4
 
 | | Crystal structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, GLYCEROL, {(3R)-6-[(3-amino-1,2,4-thiadiazol-5-yl)oxy]-1-hydroxy-4,5-dimethyl-1,3-dihydro-2,1-benzoxaborol-3-yl}acetic acid | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
4WZ5
 
 | | Crystal structure of P. aeruginosa OXA10 | | Descriptor: | Beta-lactamase OXA-10, CARBON DIOXIDE, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
Acs Infect Dis., 1, 2015
|
|
4WYY
 
 | | Crystal Structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
