4ZAR
 
 | | Crystal Structure of Proteinase K from Engyodontium albuminhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE, bound form, ... | | Authors: | Sawaya, M.R, Cascio, D, Collazo, M, Bond, C, Cohen, A, DeNicola, A, Eden, K, Jain, K, Leung, C, Lubock, N, McCormick, J, Rosinski, J, Spiegelman, L, Athar, Y, Tibrewal, N, Winter, J, Solomon, S. | | Deposit date: | 2015-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Proteinase K from Engyodontium album inhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution
to be published
|
|
6RIE
 
 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase co-transcriptional capping complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
6RID
 
 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase elongation complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
6RFL
 
 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
6RIC
 
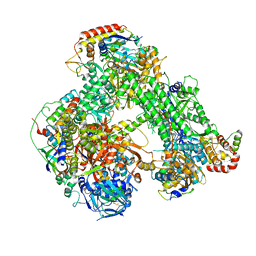 | | Structure of the core Vaccinia Virus DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, H.S, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
8U9Q
 
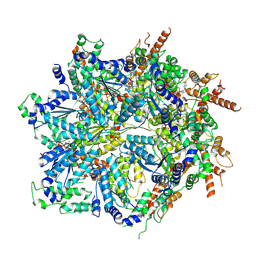 | | Cdc48-Shp1 unfolding native substrate, Class 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U9P
 
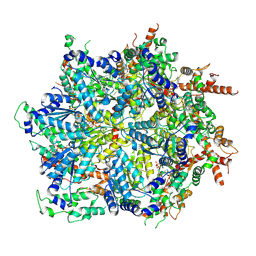 | | Cdc48-Shp1 unfolding native substrate, Class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U7T
 
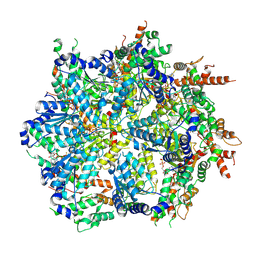 | | Substrate-bound Cdc48, Class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Lin, H.J.L, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U8I
 
 | | Cdc48-Shp1 unfolding native substrate, Class 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U9C
 
 | | Cdc48-Shp1 unfolding native substrate, Class 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8U9Z
 
 | | Cdc48-Shp1 unfolding native substrate, Class 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8UA1
 
 | | Cdc48-Shp1 unfolding native substrate, Class 9 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8UA0
 
 | | Cdc48-Shp1 unfolding native substrate, Class 8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8UAA
 
 | | Cdc48-Shp1 unfolding native substrate, Class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-20 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
8UB4
 
 | | Cdc48-Shp1 unfolding native substrate, consensus structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48, MAGNESIUM ION, ... | | Authors: | Cooney, I, Schubert, H.L, Cedeno, K, Carson, R, Fisher, O.N, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2023-09-22 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualization of the Cdc48 AAA+ ATPase protein unfolding pathway.
Nat Commun, 15, 2024
|
|
3QUX
 
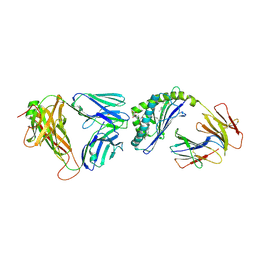 | | Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
3QUZ
 
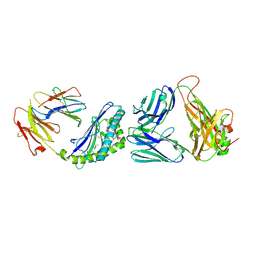 | | Structure of the mouse CD1d-NU-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
3QUY
 
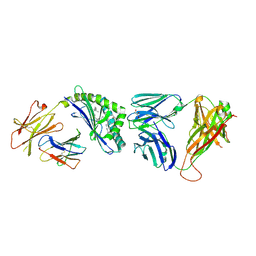 | | Structure of the mouse CD1d-BnNH-GSL-1'-iNKT TCR complex | | Descriptor: | (2S,3R,4S,5R,6S)-6-[(2S,3S,4R)-2-(hexacosanoylamino)-3,4-dihydroxy-octadecoxy]-3,4,5-trihydroxy-N-(phenylmethyl)oxane-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
8DWD
 
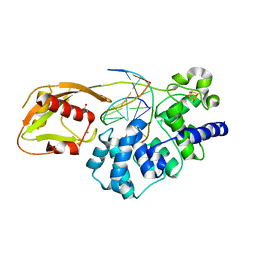 | | Adenine glycosylase MutY variant E43S in complex with DNA containing d(8-oxo-G) paired with an AP site generated by the enzyme acting on purine | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
8DWF
 
 | | Glycosylase MutY variant E43S in complex with DNA containing d(8-oxo-G) paired with substrate adenine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
8DWE
 
 | | Adenine glycosylase MutY variant E43Q in complex with DNA containing d(8-oxo-G) paired with substrate purine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Russelburg, L.P, Demir, M, David, S.S, Horvath, M.P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Base Engagement and Stereochemistry Revealed by Alteration of Catalytic Residue Glu43 in DNA Repair Glycosylase MutY
To Be Published
|
|
6RFG
 
 | |
8C8H
 
 | |
