9EVD
 
 | | In situ structure of the peripheral stalk of the mitochondrial ATPsynthase in whole Polytomella cells | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-9: Polytomella F-ATP synthase associated subunit 9, ATP synthase associated protein ASA1, ... | | Authors: | Dietrich, L, Agip, A.N.A, Kuehlbrandt, W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | In situ structure and rotary states of mitochondrial ATP synthase in whole Polytomella cells.
Science, 385, 2024
|
|
6QL4
 
 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZU
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZV
 
 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZW
 
 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZT
 
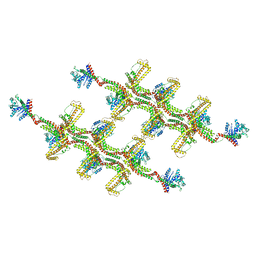 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
7B04
 
 | | Structure of Nitrite oxidoreductase (Nxr) from the anammox bacterium Kuenenia stuttgartiensis. | | Descriptor: | CALCIUM ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Moreno-Chicano, T, Dietl, A, Akram, M, Barends, T.R.M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex
Nat Microbiol, 2021
|
|
5JM4
 
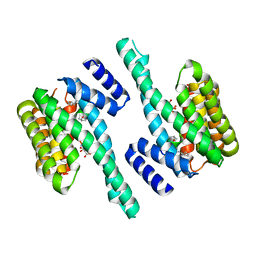 | | Crystal structure of 14-3-3zeta in complex with a cyclic peptide involving an adamantyl and a dicarboxy side chain | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, GLN-GLY-MKD-ANG-ASP-MKD-LEU-ASP-LEU-ALA-CLU | | Authors: | Bier, D, Krueger, D, Glas, A, Wallraven, K, Ottmann, C, Hennig, S, Grossmann, T. | | Deposit date: | 2016-04-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions.
J. Med. Chem., 60, 2017
|
|
