3IDE
 
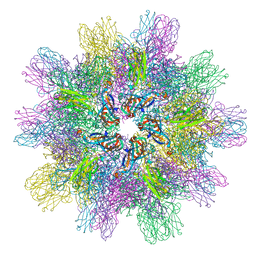 | | Structure of IPNV subviral particle | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Capsid protein VP2 | | Authors: | Coulibaly, F, Chevalier, C, Delmas, B, Rey, F.A. | | Deposit date: | 2009-07-21 | | Release date: | 2009-12-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of an aquabirnavirus particle: insights into antigenic diversity and virulence determinism
J.Virol., 84, 2010
|
|
1WCE
 
 | | Crystal structure of the T13 IBDV viral particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
1WCD
 
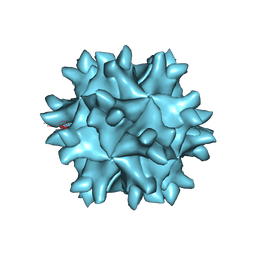 | | Crystal structure of IBDV T1 virus-like particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
1PU6
 
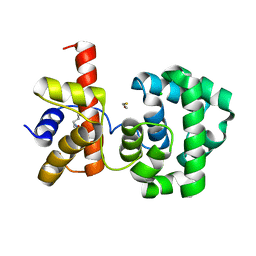 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-METHYLADENINE DNA GLYCOSYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PU7
 
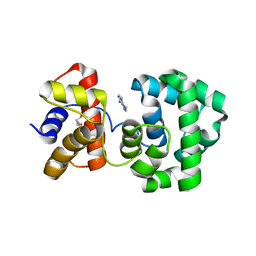 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PU8
 
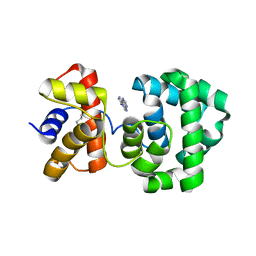 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
