4XK0
 
 | | Crystal structure of a tetramolecular RNA G-quadruplex in potassium | | Descriptor: | BARIUM ION, POTASSIUM ION, RNA (5'-(*UP*GP*GP*GP*GP*U)-3') | | Authors: | Chen, M.C, Murat, P, Abecassis, K.A, Ferre-D'Amare, A.R, Balasubramanian, S. | | Deposit date: | 2015-01-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase.
Nucleic Acids Res., 43, 2015
|
|
4LCQ
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBI | | Descriptor: | (2S)-3-(carbamoylamino)-2-methylpropanoic acid, ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4LCR
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBA | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4LCS
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with hydantoin | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, ZINC ION, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S. | | Deposit date: | 2013-06-23 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4TS2
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, magnesium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4TS0
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, barium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, BARIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4H00
 
 | | The crystal structure of mon-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4GZ7
 
 | | The crystal structure of Apo-dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, dihydropyrimidinase | | Authors: | Hsien, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structure Requirements for Post-translational Modification
To be Published
|
|
4H01
 
 | | The crystal structure of di-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4OX5
 
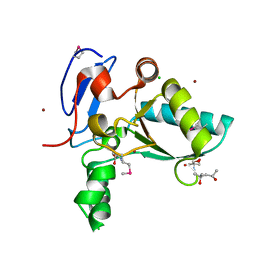 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OXD
 
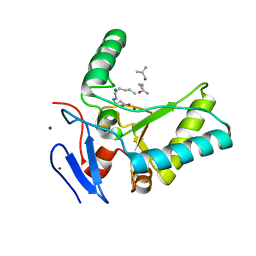 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | CHLORIDE ION, LYSINE, LdcB LD-carboxypeptidase, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OX3
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | PHOSPHATE ION, Putative carboxypeptidase YodJ, ZINC ION | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
5VHE
 
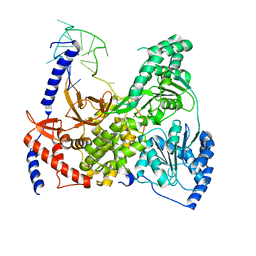 | | DHX36 in complex with the c-Myc G-quadruplex | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36, DNA (5'-D(*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*TP*TP*TP*TP*T)-3'), POTASSIUM ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.793 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHD
 
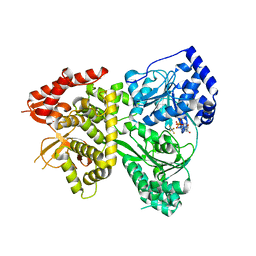 | | DHX36 with an N-terminal truncation bound to ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEAH (Asp-Glu-Ala-His) box polypeptide 36, TETRAFLUOROALUMINATE ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHC
 
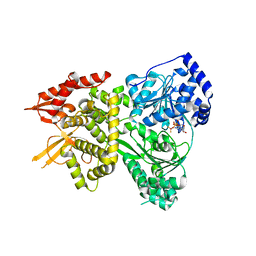 | | DHX36 with an N-terminal truncation bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DEAH (Asp-Glu-Ala-His) box polypeptide 36, ... | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHA
 
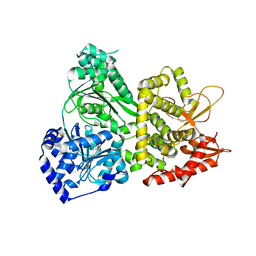 | | DHX36 with an N-terminal truncation | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
7Y6W
 
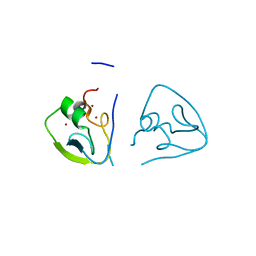 | | RRGSGG-AtPRT6 UBR box (I222) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6Z
 
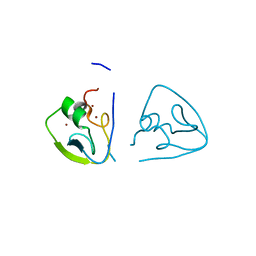 | | RLGSGG-AtPRT6 UBR box (I222) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWG
 
 | | RSGSGG-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWE
 
 | | RRGSGG-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, MAGNESIUM ION, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWF
 
 | | RLGSGG-AtPRT6 UBR box (highest resolution) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y70
 
 | | RLGSGG-AtPRT6 UBR box (P4332) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWD
 
 | | Apo-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6Y
 
 | | RLGSGG-AtPRT6 UBR box (C121) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6X
 
 | | RRGSGG-AtPRT6 UBR box (P32) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
