2ACO
 
 | | Xray structure of Blc dimer in complex with vaccenic acid | | Descriptor: | Outer membrane lipoprotein blc, VACCENIC ACID | | Authors: | Campanacci, V, Bishop, R.E, Reese, L, Blangy, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2005-07-19 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The membrane bound bacterial lipocalin Blc is a functional dimer with binding preference for lysophospholipids.
Febs Lett., 580, 2006
|
|
1QWD
 
 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | Deposit date: | 2003-09-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
7Q1E
 
 | | CPAP:TUBULIN:IIH5 ALPHAREP COMPLEX | | Descriptor: | Centromere protein J, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Campanacci, V, Gigant, b. | | Deposit date: | 2021-10-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q1F
 
 | | CPAP:TUBULIN:IE5 ALPHAREP COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CITRATE ANION, Centromere protein J, ... | | Authors: | Campanacci, V, Gigant, b. | | Deposit date: | 2021-10-19 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4BET
 
 | | Crystal structure of the Legionella pneumophila FIC domain-containing effector AnkX protein (inactive H229A mutant) in complex with cytidine-diphosphate-choline | | Descriptor: | GLYCEROL, PHOSPHOCHOLINE TRANSFERASE ANKX, SULFATE ION, ... | | Authors: | Campanacci, V, Mukherjee, S, Roy, C.R, Cherfils, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the Legionella Effector Ankx Reveals the Mechanism of Phosphocholine Transfer by the Fic Domain.
Embo J., 32, 2013
|
|
4BER
 
 | | Crystal structure of the Legionella pneumophila FIC domain-containing effector AnkX protein in complex with cytidine monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Campanacci, V, Mukherjee, S, Roy, C.R, Cherfils, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Legionella Effector Ankx Reveals the Mechanism of Phosphocholine Transfer by the Fic Domain.
Embo J., 32, 2013
|
|
4BEP
 
 | | Crystal structure of the Legionella pneumophila FIC domain-containing effector AnkX protein (apo-form) | | Descriptor: | MAGNESIUM ION, PHOSPHOCHOLINE TRANSFERASE ANKX, SULFATE ION | | Authors: | Campanacci, V, Mukherjee, S, Roy, C.R, Cherfils, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of the Legionella Effector Ankx Reveals the Mechanism of Phosphocholine Transfer by the Fic Domain.
Embo J., 32, 2013
|
|
4BES
 
 | | Crystal structure of the Legionella pneumophila FIC domain-containing effector AnkX protein in complex with cytidine monophosphate and phosphocholine | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, PHOSPHOCHOLINE, PHOSPHOCHOLINE TRANSFERASE ANKX, ... | | Authors: | Campanacci, V, Mukherjee, S, Roy, C.R, Cherfils, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of the Legionella Effector Ankx Reveals the Mechanism of Phosphocholine Transfer by the Fic Domain.
Embo J., 32, 2013
|
|
1NZJ
 
 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | Descriptor: | Hypothetical protein yadB, ZINC ION | | Authors: | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | Deposit date: | 2003-02-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
1N8V
 
 | | Chemosensory Protein in complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1N8U
 
 | | Chemosensory Protein in Complex with bromo-dodecanol | | Descriptor: | BROMO-DODECANOL, chemosensory protein | | Authors: | Campanacci, V, Lartigue, A, Hallberg, B.M, Jones, T.A, Giudici-Orticoni, M.T, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-11-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7Z0G
 
 | | CPAP:TUBULIN:IE5 ALPHAREP COMPLEX P1 SPACE GROUP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Centromere protein J, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z0F
 
 | | CPAP:S-TUBULIN:IIH5 ALPHAREP COMPLEX | | Descriptor: | Centromere protein J, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural convergence for tubulin binding of CPAP and vinca domain microtubule inhibitors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6GX7
 
 | | Tubulin-CopN-alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-26 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GWC
 
 | | Tubulin:iE5 alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALPHA-TUBULIN, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-22 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Structure, 27, 2019
|
|
6GVN
 
 | | Tubulin:TM-3 DARPin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Cantos Fernandes, S, Campanacci, V. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GWD
 
 | | Tubulin:iiH5 alphaRep complex | | Descriptor: | ALPHA-TUBULIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-22 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Structure, 27, 2019
|
|
6GVM
 
 | | Tubulin:F3II DARPin complex | | Descriptor: | F3II DARPIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V, Cantos Fernandes, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1KX9
 
 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
1KX8
 
 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
1QZ8
 
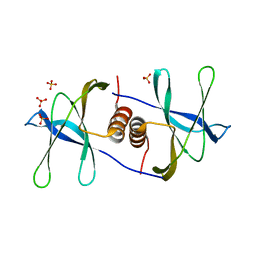 | | Crystal structure of SARS coronavirus NSP9 | | Descriptor: | SULFATE ION, polyprotein 1ab | | Authors: | Egloff, M.P, Ferron, F, Campanacci, V, Longhi, S, Rancurel, C, Dutartre, H, Snijder, E.J, Gorbalenya, A.E, Cambillau, C, Canard, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4V5I
 
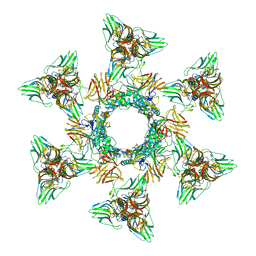 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1U08
 
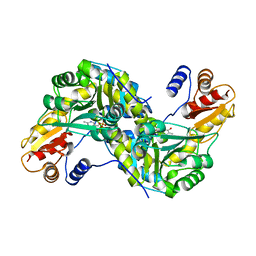 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
1QYA
 
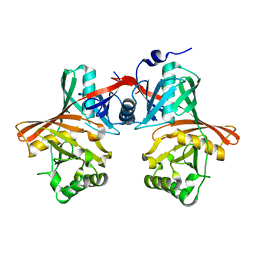 | | CRYSTAL STRUCTURE OF E. COLI PROTEIN YDDE | | Descriptor: | HYPOTHETICAL PROTEIN yddE | | Authors: | Grassick, A, Sulzenbacher, G, Roig-Zamboni, V, Campanacci, V, Cambillau, C, Bourne, Y. | | Deposit date: | 2003-09-10 | | Release date: | 2004-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of E. coli yddE protein reveals a striking homology with diaminopimelate epimerase
Proteins, 55, 2004
|
|
1QY9
 
 | | Crystal structure of E. coli Se-MET protein YDDE | | Descriptor: | GLYCEROL, HYDROXIDE ION, HYPOTHETICAL PROTEIN yddE | | Authors: | Grassick, A, Sulzenbacher, G, Roig-Zamboni, V, Campanacci, V, Cambillau, C, Bourne, Y. | | Deposit date: | 2003-09-10 | | Release date: | 2004-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of E. coli yddE protein reveals a striking homology with diaminopimelate epimerase
Proteins, 55, 2004
|
|
