1P4U
 
 | | CRYSTAL STRUCTURE OF GGA3 GAE DOMAIN IN COMPLEX WITH RABAPTIN-5 PEPTIDE | | Descriptor: | ADP-ribosylation factor binding protein GGA3, Rabaptin-5 | | Authors: | Miller, G.J, Mattera, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RECOGNITION OF ACCESSORY PROTEIN MOTIFS BY THE GAMMA-ADAPTIN EAR DOMAIN OF GGA3
Nat.Struct.Biol., 10, 2003
|
|
4HMY
 
 | | Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Ren, X, Farias, G.G, Canagarajah, B.J, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2012-10-18 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structural Basis for Recruitment and Activation of the AP-1 Clathrin Adaptor Complex by Arf1.
Cell(Cambridge,Mass.), 152, 2013
|
|
6WR4
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
6WQZ
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
8AFZ
 
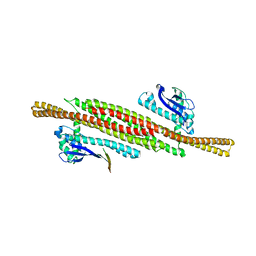 | | Architecture of the ESCPE-1 membrane coat | | Descriptor: | Cation-independent mannose-6-phosphate receptor, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E, Ishida, M, Williamsom, C.D, Banos-Mateos, S, Gil-Carton, D, Romero, M, Vidaurrazaga, A, Fernandez-Recio, J, Rojas, A.L, Bonifacino, J.S, Castano-Diez, D, Hierro, A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3L81
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain, in complex with a sorting peptide from the amyloid precursor protein (APP) | | Descriptor: | AP-4 complex subunit mu-1, Amyloid beta A4 protein, GLYCEROL | | Authors: | Mardones, G.A, Rojas, A.L, Burgos, P.V, Dasilva, L.L.P, Prabhu, Y, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2009-12-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex.
Dev.Cell, 18, 2010
|
|
3N1B
 
 | | C-terminal domain of Vps54 subunit of the GARP complex | | Descriptor: | Vacuolar protein sorting-associated protein 54 | | Authors: | Perez-Victoria, F.J, Abascal-Palacios, G, Tascon, I, Kajava, A, Pioro, E.P, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2010-05-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3N1E
 
 | | Vps54 C-terminal domain | | Descriptor: | GLYCEROL, MAGNESIUM ION, Vacuolar protein sorting-associated protein 54 | | Authors: | Perez-Victoria, F.J, Abascal-Palacios, G, Tascon, I, Kajava, A, Pioro, E.P, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2010-05-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1YD8
 
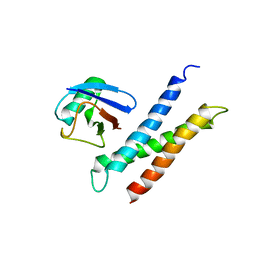 | | COMPLEX OF HUMAN GGA3 GAT DOMAIN AND UBIQUITIN | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, UBIQUIN | | Authors: | Prag, G, Lee, S, Mattera, R, Arighi, C.N, Beach, B.M, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, {gamma}-ear-containing, ADP-ribosylation-factor-binding proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1LF8
 
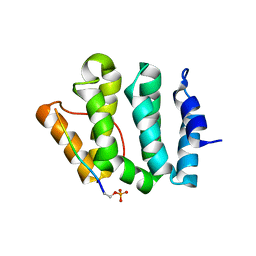 | | Complex of GGA3-VHS Domain and CI-MPR C-terminal Phosphopeptide | | Descriptor: | ADP-ribosylation factor binding protein GGA3, Cation-independent mannose-6-phosphate receptor | | Authors: | Kato, Y, Misra, S, Puertollano, R, Hurley, J.H, Bonifacino, J.S. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism.
Nat.Struct.Biol., 9, 2002
|
|
1JUQ
 
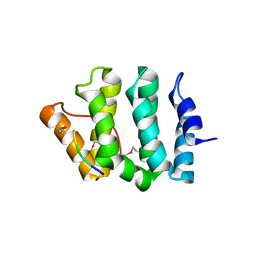 | | GGA3 VHS domain complexed with C-terminal peptide from cation-dependent Mannose 6-phosphate receptor | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, Cation-dependent mannose-6-phosphate receptor | | Authors: | Misra, S, Puertollano, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2001-08-26 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains.
Nature, 415, 2002
|
|
1JPL
 
 | | GGA3 VHS domain complexed with C-terminal peptide from cation-independent mannose 6-phosphate receptor | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, Cation-Independent Mannose 6-phosphate receptor | | Authors: | Misra, S, Puertollano, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2001-08-02 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains.
Nature, 415, 2002
|
|
4NEE
 
 | | crystal structure of AP-2 alpha/simga2 complex bound to HIV-1 Nef | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit sigma, Protein Nef | | Authors: | Hurley, J.H, Bonifacino, J.S, Ren, X, Park, S.Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8841 Å) | | Cite: | How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4.
Elife, 3, 2014
|
|
4IKN
 
 | | Crystal structure of adaptor protein complex 3 (AP-3) mu3A subunit C-terminal domain, in complex with a sorting peptide from TGN38 | | Descriptor: | AP-3 complex subunit mu-1, Trans-Golgi network integral membrane protein TGN38 | | Authors: | Mardones, G.A, Kloer, D.P, Burgos, P.V, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2012-12-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for the recognition of tyrosine-based sorting signals by the mu 3A subunit of the AP-3 adaptor complex.
J.Biol.Chem., 288, 2013
|
|
4J2C
 
 | | GARP-SNARE Interaction | | Descriptor: | Syntaxin-6, Vacuolar protein sorting-associated protein 51 homolog | | Authors: | Abascal-Palacios, G, Schindler, C, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the interaction of the Golgi-Associated Retrograde Protein Complex with the t-SNARE Syntaxin 6.
Structure, 21, 2013
|
|
2R17
 
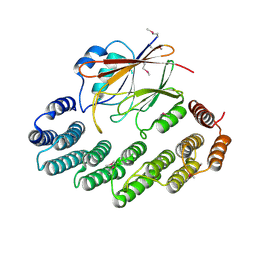 | | Functional architecture of the retromer cargo-recognition complex | | Descriptor: | GLYCEROL, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Hierro, A, Rojas, A.L, Rojas, R, Murthy, N, Effantin, G, Kajava, A.V, Steven, A.C, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2007-08-22 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional architecture of the retromer cargo-recognition complex.
Nature, 449, 2007
|
|
2FAU
 
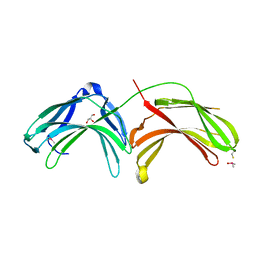 | | Crystal structure of human vps26 | | Descriptor: | GLYCEROL, Vacuolar protein sorting 26 | | Authors: | Shi, H, Rojas, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5F0P
 
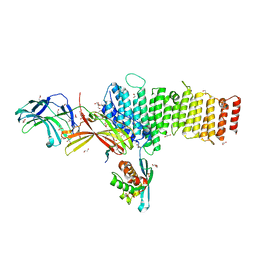 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1(L557M) (SeMet labeled) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5F0J
 
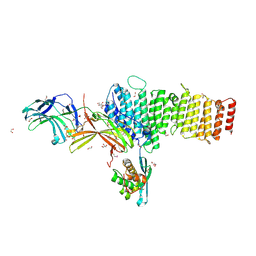 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5F0L
 
 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5F0K
 
 | | Structure of VPS35 N terminal region | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.074 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5F0M
 
 | | Structure of retromer VPS26-VPS35 subunits bound to SNX3 and DMT1 (SeMet labeled) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Natural resistance-associated macrophage protein 2, ... | | Authors: | Lucas, M, Gershlick, D, Vidaurrazaga, A, Rojas, A.L, Bonifacino, J.S, Hierro, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Mechanism for Cargo Recognition by the Retromer Complex.
Cell, 167, 2016
|
|
5OT4
 
 | | Structure of the Legionella pneumophila effector RidL (1-866) | | Descriptor: | GLYCEROL, Interaptin | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OSI
 
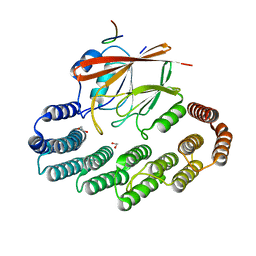 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL harpin loop (163-176) | | Descriptor: | 1,2-ETHANEDIOL, Interaptin, SODIUM ION, ... | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OSH
 
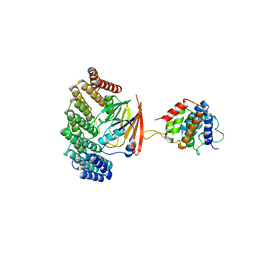 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL N-terminal domain (1-236) | | Descriptor: | Interaptin, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
