6SYF
 
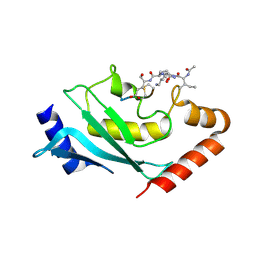 | | Human Ubc9 with covalent isopeptide ligand | | Descriptor: | ACE-ILE-LYS-GLN-GLU, ACE-LEU-ARG-LEU-ARG-GLY-CYS, SUMO-conjugating enzyme UBC9 | | Authors: | Hofmann, R, Akimoto, G, Wucherpfennig, T.G, Zeymer, C, Bode, J.W. | | Deposit date: | 2019-09-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine acylation using conjugating enzymes for site-specific modification and ubiquitination of recombinant proteins.
Nat.Chem., 12, 2020
|
|
8AFI
 
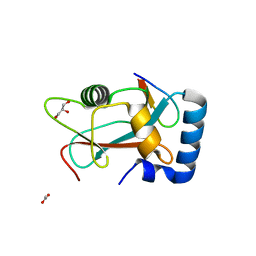 | | GABARAP in complex with LIR motif of HsATG3 | | Descriptor: | ACETATE ION, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Farnung, J, Benoit, R.M, Corn, J.E, Bode, J.W. | | Deposit date: | 2022-07-18 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Semisynthetic LC3 Probes for Autophagy Pathways Reveal a Noncanonical LC3 Interacting Region Motif Crucial for the Enzymatic Activity of Human ATG3.
Acs Cent.Sci., 9, 2023
|
|
9FMM
 
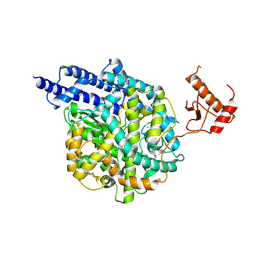 | | Structure of human ACE2 in complex with a fluorinated small molecule inhibitor | | Descriptor: | (2~{S})-2-[[(2~{S})-3-[3-[(3-chloranyl-5-fluoranyl-phenyl)methyl]imidazol-4-yl]-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-4-methyl-pentanoic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Benoit, R.B, Rodrigues, M.J, Wieser, M.M. | | Deposit date: | 2024-06-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of radiofluorinated MLN-4760 derivatives for PET imaging of the SARS-CoV-2 entry receptor ACE2.
Eur J Nucl Med Mol Imaging, 2024
|
|
7O7Z
 
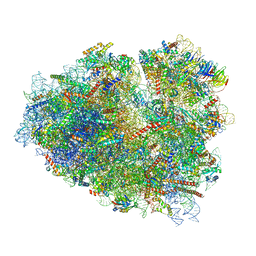 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
