6YZ5
 
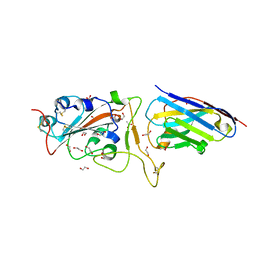 | | H11-D4 complex with SARS-CoV-2 RBD | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, Le Bas, A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
6YZ7
 
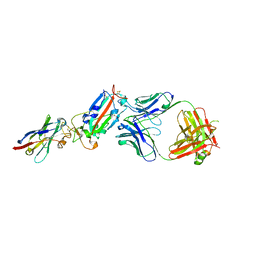 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
6Z2M
 
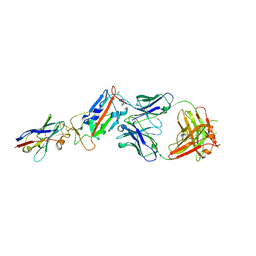 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 antibody, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published
|
|
5CNA
 
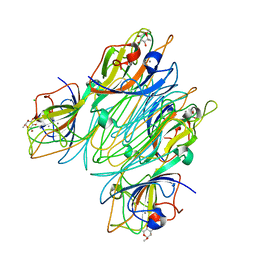 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
6ZBP
 
 | | H11-H4 complex with SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H11-H4, SULFATE ION, ... | | Authors: | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, LeBas, A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
7AN4
 
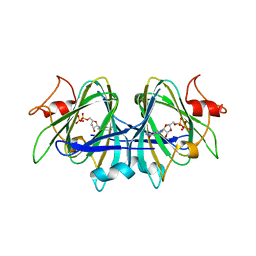 | |
7ANJ
 
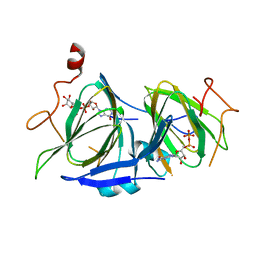 | |
7ANH
 
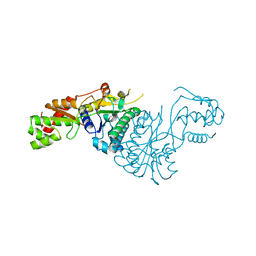 | | DdhaC | | Descriptor: | GDP-L-fucose synthase | | Authors: | Naismith, J.H, Woodward, L. | | Deposit date: | 2020-10-11 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | DdhaC
To Be Published
|
|
7ANI
 
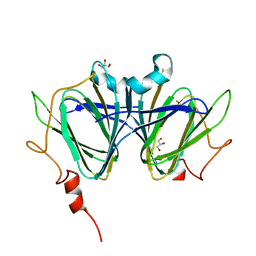 | |
7ANG
 
 | |
7ANC
 
 | |
1CVN
 
 | | CONCANAVALIN A COMPLEXED TO TRIMANNOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Naismith, J.H. | | Deposit date: | 1995-08-09 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of trimannoside recognition by concanavalin A.
J.Biol.Chem., 271, 1996
|
|
1EXT
 
 | |
1NCF
 
 | |
1CON
 
 | | THE REFINED STRUCTURE OF CADMIUM SUBSTITUTED CONCANAVALIN A AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CADMIUM ION, CALCIUM ION, CONCANAVALIN A | | Authors: | Naismith, J.H, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Kalb(Gilboa), A.J, Yariv, J, Wan, T.C.M, Weisgerber, S. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of cadmium-substituted concanavalin A at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
6ENK
 
 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
6Z3C
 
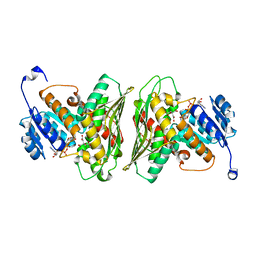 | | High resolution structure of RgNanOx | | Descriptor: | CITRATE ANION, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M.O. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6Z33
 
 | |
6Z3B
 
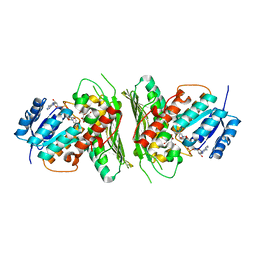 | | Low resolution structure of RgNanOx | | Descriptor: | CITRIC ACID, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6Z2N
 
 | |
6YY5
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with TCV_L5 | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[2-[[(2~{S})-3-[[(2~{S})-3-[[1-[2-[2-[2-[4-[4-[5-(acetamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]-2-fluoranyl-phenyl]piperazin-1-yl]-2-oxidanylidene-ethoxy]ethoxy]ethyl]-1,2,3-triazol-4-yl]methylamino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-oxidanylidene-ethyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Naismith, J.H, Moynie, L.M. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Hijacking of the Enterobactin Pathway by a Synthetic Catechol Vector Designed for Oxazolidinone Antibiotic Delivery in Pseudomonas aeruginosa.
Acs Infect Dis., 2022
|
|
7OAO
 
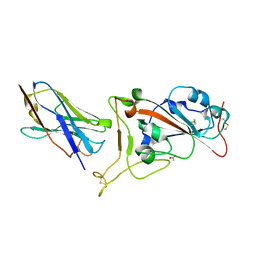 | | Nanobody C5 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAQ
 
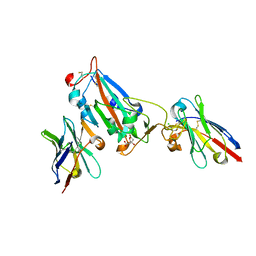 | | Nanobody H3 AND C1 bound to RBD with Kent mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
