5YOB
 
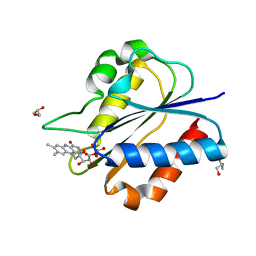 | | Crystal Structure of flavodoxin without engineered disulfide bond | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Pu, M, Xu, Z, Song, G, Liu, Z.J. | | Deposit date: | 2017-10-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.142 Å) | | Cite: | Protein crystal quality oriented disulfide bond engineering.
Protein Cell, 9, 2018
|
|
5YT4
 
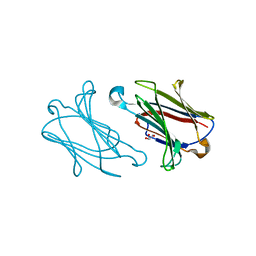 | | Galectin-10 variant H53A soaked in glycerol for 5 minutes | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5HW5
 
 | |
8JM9
 
 | |
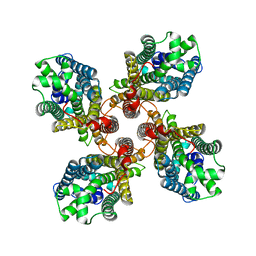
8JMI
 
 | |
8JMA
 
 | |
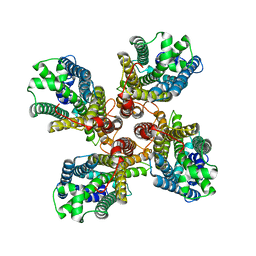
8JME
 
 | |
8JMH
 
 | |
6A3N
 
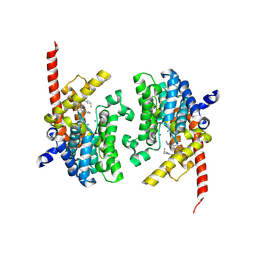 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 2 | | Descriptor: | 1-cyclopentyl-6-({(2R)-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxopropan-2-yl}amino)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y.N, Zhou, Q, Chen, Y.P, Luo, H.B. | | Deposit date: | 2018-06-15 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors against Phosphodiesterase-9, a Novel Target for the Treatment of Vascular Dementia.
J. Med. Chem., 62, 2019
|
|
5HVI
 
 | |
5ZUU
 
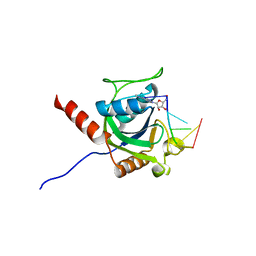 | | Crystal structure of AtCPSF30 YTH domain in complex with 10mer m6A-modified RNA | | Descriptor: | 30-kDa cleavage and polyadenylation specificity factor 30, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Nie, H, Li, S, Patel, D.J. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis.
Mol Plant, 2021
|
|
5HW1
 
 | |
7XFR
 
 | |
8K0B
 
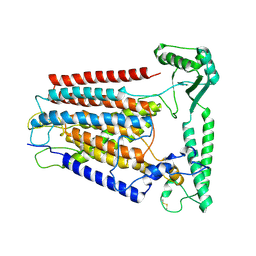 | | Cryo-EM structure of TMEM63C | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Qin, Y, Yu, D, Dong, J, Dang, S. | | Deposit date: | 2023-07-08 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure of TMEM63C suggests it functions as a monomer.
Nat Commun, 14, 2023
|
|
5IQ8
 
 | |
6JEA
 
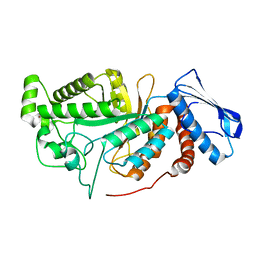 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6IPQ
 
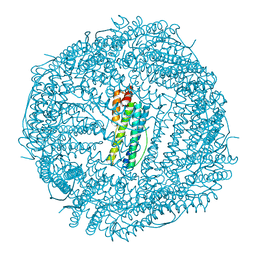 | | Non-native ferritin 8-mer mutant-C90A/C102A/C130A | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION | | Authors: | Zang, J, Chen, H, Wang, Y, Zhao, G. | | Deposit date: | 2018-11-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Disulfide-mediated conversion of 8-mer bowl-like protein architecture into three different nanocages.
Nat Commun, 10, 2019
|
|
5I52
 
 | |
5I63
 
 | |
8HBC
 
 | | Crystal structure of the CysR-CTLD3 fragment of human DEC205 | | Descriptor: | Lymphocyte antigen 75 | | Authors: | Kong, D, Yu, B, Hu, Z, Cheng, C, Cao, L, He, Y. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Interaction of human dendritic cell receptor DEC205/CD205 with keratins.
J.Biol.Chem., 300, 2024
|
|
1HZI
 
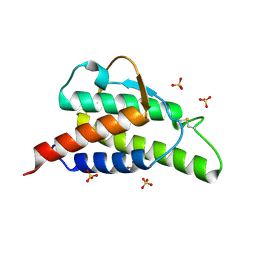 | | INTERLEUKIN-4 MUTANT E9A | | Descriptor: | INTERLEUKIN-4, SULFATE ION | | Authors: | Hulsmeyer, M, Scheufler, C, Dreyer, M.K. | | Deposit date: | 2001-01-25 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of interleukin 4 mutant E9A suggests polar steering in receptor-complex formation.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7WK2
 
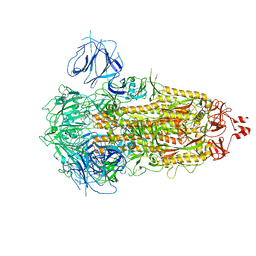 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WKA
 
 | |
7WK9
 
 | |
7WK8
 
 | |
