3VOA
 
 | | Staphylococcus aureus FtsZ 12-316 GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2YVE
 
 | | Crystal structure of the methylene blue-bound form of the multi-drug binding transcriptional repressor CgmR | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, GLYCEROL, ... | | Authors: | Itou, H, Shirakihara, Y, Tanaka, I. | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
3VO9
 
 | | Staphylococcus aureus FtsZ apo-form (SeMet) | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2YVH
 
 | |
2Z2F
 
 | | X-ray Crystal Structure of Bovine Stomach Lysozyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme C-2, SODIUM ION | | Authors: | Akieda, D, Nonaka, Y, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability of Bovine Stomach Lysozyme in Acidic Condition
To be Published
|
|
2Z2E
 
 | | Crystal Structure of Canine Milk Lysozyme Stabilized against Non-enzymatic Deamidation | | Descriptor: | Lysozyme C, milk isozyme, SULFATE ION | | Authors: | Nonaka, Y, Akieda, D, Watanabe, N, Tanaka, I, Kamiya, M, Aizawa, T, Nitta, K, Demura, M, Kawano, K. | | Deposit date: | 2007-05-21 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Spontaneous asparaginyl deamidation of canine milk lysozyme under mild conditions
Proteins, 72, 2008
|
|
3VO8
 
 | | Staphylococcus aureus FtsZ GDP-form | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Matsui, T, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VPA
 
 | | Staphylococcus aureus FtsZ apo-form | | Descriptor: | Cell division protein FtsZ | | Authors: | Matsui, T, Yamane, J, Mogi, N, Yao, M, Tanaka, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ZOY
 
 | | The multi-drug binding transcriptional repressor CgmR (CGL2612 protein) from C.glutamicum | | Descriptor: | GLYCEROL, Transcriptional regulator | | Authors: | Itou, H, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The CGL2612 protein from Corynebacterium glutamicum is a drug resistance-related transcriptional repressor: structural and functional analysis of a newly identified transcription factor from genomic DNA analysis
J.Biol.Chem., 280, 2005
|
|
2ZCX
 
 | |
3VOB
 
 | | Staphylococcus aureus FtsZ with PC190723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Yamane, J, Matsui, T, Mogi, N, Yamaguchi, H, Takemoto, H, Yao, M, Tanaka, I. | | Deposit date: | 2012-01-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ZU7
 
 | |
3VW5
 
 | |
3VXF
 
 | | X/N Joint refinement of Human alpha-thrombin-Bivalirudin complex PD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.602 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
2ZPA
 
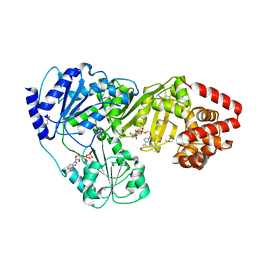 | | Crystal Structure of tRNA(Met) Cytidine Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Chimnaronk, S, Manita, T, Yao, M, Tanaka, I. | | Deposit date: | 2008-07-08 | | Release date: | 2009-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon
Embo J., 28, 2009
|
|
2ZDS
 
 | | Crystal Structure of SCO6571 from Streptomyces coelicolor A3(2) | | Descriptor: | Putative DNA-binding protein | | Authors: | Begum, P, Gao, Y.G, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-02 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SCO6571 from Streptomyces coelicolor A3(2).
Protein Pept.Lett., 15, 2008
|
|
2ZZF
 
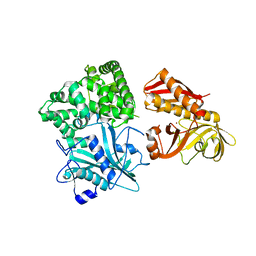 | | Crystal structure of alanyl-tRNA synthetase without oligomerization domain | | Descriptor: | Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3VI6
 
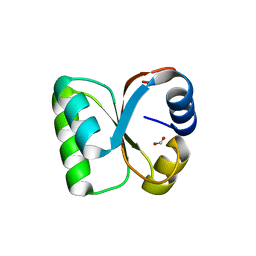 | | Crystal Structure of human ribosomal protein L30e | | Descriptor: | 60S ribosomal protein L30, FORMIC ACID | | Authors: | Kawaguchi, A, Ose, T, Yao, M, Tanaka, I. | | Deposit date: | 2011-09-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallization and preliminary X-ray structure analysis of human ribosomal protein L30e
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3A1R
 
 | |
3A7W
 
 | |
3A88
 
 | |
3A7V
 
 | |
3A87
 
 | |
3A1Y
 
 | | The structure of archaeal ribosomal stalk P1/P0 complex | | Descriptor: | 50S ribosomal protein P1 (L12P), Acidic ribosomal protein P0 | | Authors: | Naganuma, T, Yao, M, Nomura, N, Yu, J, Uchiumi, T, Tanaka, I. | | Deposit date: | 2009-04-25 | | Release date: | 2009-11-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for translation factor recruitment to the eukaryotic/archaeal ribosomes
J.Biol.Chem., 285, 2010
|
|
3VXE
 
 | | Human alpha-thrombin-Bivalirudin complex at PD5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIVALIRUDIN, Thrombin heavy chain, ... | | Authors: | Yamada, T, Kurihara, K, Masumi, K, Tamada, T, Tomoyori, K, Ohnishi, Y, Tanaka, I, Kuroki, R, Niimura, N. | | Deposit date: | 2012-09-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Neutron and X-ray crystallographic analysis of the human alpha-thrombin-bivalirudin complex at pD 5.0: protonation states and hydration structure of the enzyme-product complex
Biochim.Biophys.Acta, 1834, 2013
|
|
