3OW3
 
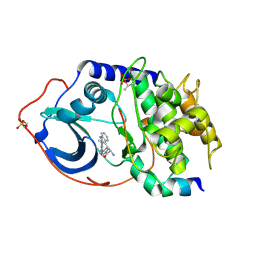 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW4
 
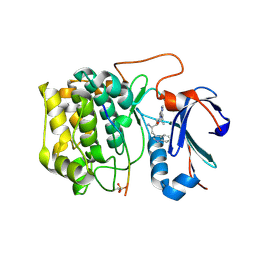 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, GSK 3 beta peptide, RAC-alpha serine/threonine-protein kinase | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7ZUN
 
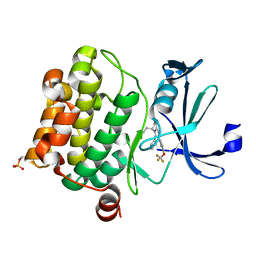 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone compound | | Descriptor: | (4~{S})-4-(2-azanylethyl)-6-phenyl-7-[3-(trifluoromethyloxy)phenyl]-3,4-dihydropyrrolo[1,2-a]pyrazin-1-ol, Isoform 2 of Serine/threonine-protein kinase pim-1 | | Authors: | Casale, E. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereoselective synthesis of 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one derivatives as PIM kinase inhibitors inspired from marine alkaloids.
Chirality, 34, 2022
|
|
8AUD
 
 | |
8AU6
 
 | |
4D7W
 
 | | Crystal structure of sortase C1 (SrtC1) from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, SORTASE FAMILY PROTEIN | | Authors: | Malito, E, Lazzarin, M, Cozzi, R, Bottomley, M.J. | | Deposit date: | 2014-11-28 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Noncanonical Sortase-Mediated Assembly of Pilus Type 2B in Group B Streptococcus.
Faseb J., 29, 2015
|
|
2K3J
 
 | | The solution structure of human Mia40 | | Descriptor: | Mitochondrial intermembrane space import and assembly protein 40 | | Authors: | Ciofi Baffoni, S, Bertini, I, Gallo, A. | | Deposit date: | 2008-05-08 | | Release date: | 2009-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria.
Nat.Struct.Mol.Biol., 16, 2009
|
|
