4J69
 
 | | Crystal structure of Ribonuclease A soaked in 50% S,R,S-bisfuranol: One of twelve in MSCS set | | Descriptor: | (3S,3aR,6aS)-hexahydrofuro[2,3-b]furan-3-ol, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4J6A
 
 | | Crystal structure of Ribonuclease A soaked in 40% 2,2,2-Trifluoroethanol: One of twelve in MSCS set | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, TRIFLUOROETHANOL | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4J63
 
 | |
4J61
 
 | |
4J66
 
 | |
4J68
 
 | |
4J5Z
 
 | |
4J65
 
 | |
4J67
 
 | | Crystal structure of Ribonuclease A soaked in 50% 1,6-Hexanediol: One of twelve in MSCS set | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4J60
 
 | |
4J64
 
 | | Crystal structure of Ribonuclease A soaked in 40% Dioxane: One of twelve in MSCS set | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
1D8F
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A PIPERAZINE BASED INHIBITOR. | | Descriptor: | CALCIUM ION, N-HYDROXY-1-(4-METHOXYPHENYL)SULFONYL-4-BENZYLOXYCARBONYL-PIPERAZINE-2-CARBOXAMIDE, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Cheng, M.Y, De, B, Pikul, S, Almstead, N.G, Natchus, M.G, Anastasio, M.V, McPhail, S.J, Snider, C.E, Taiwo, Y.O, Chen, L.Y. | | Deposit date: | 1999-10-22 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of piperazine-based matrix metalloproteinase inhibitors.
J.Med.Chem., 43, 2000
|
|
5BV9
 
 | |
6L30
 
 | | Crystal structure of the epithelial cell transforming 2 (ECT2) | | Descriptor: | Protein ECT2 | | Authors: | Chen, Z.C, Chen, M.R, Pan, H, Sun, L.F, Shi, P. | | Deposit date: | 2019-10-07 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and regulation of human epithelial cell transforming 2 protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5VHE
 
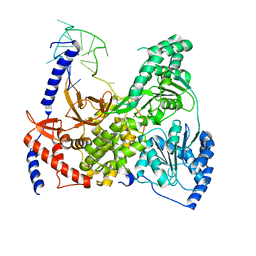 | | DHX36 in complex with the c-Myc G-quadruplex | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36, DNA (5'-D(*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*TP*TP*TP*TP*T)-3'), POTASSIUM ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.793 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHD
 
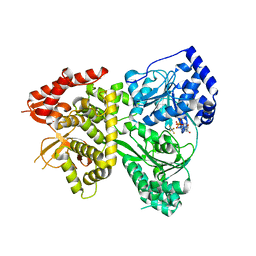 | | DHX36 with an N-terminal truncation bound to ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEAH (Asp-Glu-Ala-His) box polypeptide 36, TETRAFLUOROALUMINATE ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHC
 
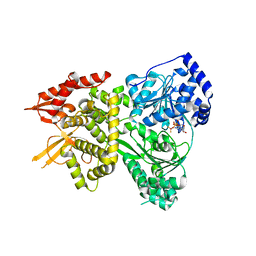 | | DHX36 with an N-terminal truncation bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DEAH (Asp-Glu-Ala-His) box polypeptide 36, ... | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHA
 
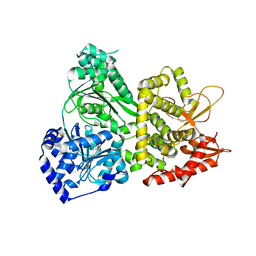 | | DHX36 with an N-terminal truncation | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
7CMC
 
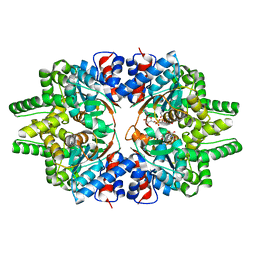 | | CRYSTAL STRUCTURE OF DEOXYHYPUSINE SYNTHASE FROM PYROCOCCUS HORIKOSHII | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable deoxyhypusine synthase | | Authors: | Yu, J, Gai, Z.Q, Okada, C, Yao, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexible NAD+Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A.
Int J Mol Sci, 21, 2020
|
|
1D7X
 
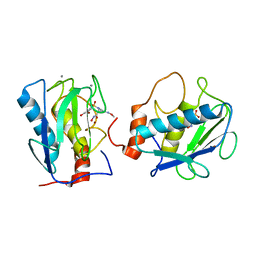 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A MODIFIED PROLINE SCAFFOLD BASED INHIBITOR. | | Descriptor: | CALCIUM ION, N-HYDROXY 1N(4-METHOXYPHENYL)SULFONYL-4-(Z,E-N-METHOXYIMINO)PYRROLIDINE-2R-CARBOXAMIDE, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Cheng, M.Y, Natchus, M.G, De, B, Almstead, N.G, Pikul, S. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of matrix metalloproteinase inhibitors derived from a modified proline scaffold.
J.Med.Chem., 42, 1999
|
|
5C6N
 
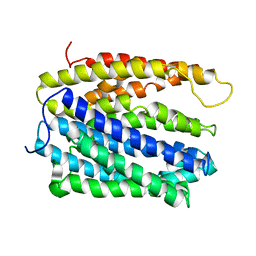 | | protein A | | Descriptor: | BH2163 protein | | Authors: | Lu, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the blockade of MATE multidrug efflux pumps.
Nat Commun, 6, 2015
|
|
5C6P
 
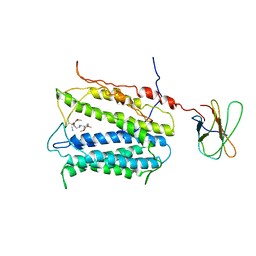 | | protein C | | Descriptor: | (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](methyl)amino}-2-(propan-2-yl)pentanenitrile, protein C, protein D | | Authors: | Lu, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the blockade of MATE multidrug efflux pumps.
Nat Commun, 6, 2015
|
|
5C6O
 
 | | protein B | | Descriptor: | (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](methyl)amino}-2-(propan-2-yl)pentanenitrile, BH2163 protein | | Authors: | Lu, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the blockade of MATE multidrug efflux pumps.
Nat Commun, 6, 2015
|
|
7YTJ
 
 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
4ZPG
 
 | |
