2YUR
 
 | | Solution structure of the Ring finger of human Retinoblastoma-binding protein 6 | | Descriptor: | Retinoblastoma-binding protein 6, ZINC ION | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Tomizawa, T, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ring finger of human Retinoblastoma-binding protein 6
To be Published
|
|
7EMM
 
 | | Crystal structure of IrCp* immobilized apo-R52H-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Taher, M, Maity, B, Nakane, T, Abe, S, Ueno, T, Mazumdar, S. | | Deposit date: | 2021-04-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Controlled Uptake of an Iridium Complex inside Engineered apo-Ferritin Nanocages: Study of Structure and Catalysis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7EML
 
 | | Structure of IrCp* immobilized apo-D38H-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Taher, M, Maity, B, Nakane, T, Abe, S, Ueno, T, Mazumdar, S. | | Deposit date: | 2021-04-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Controlled Uptake of an Iridium Complex inside Engineered apo-Ferritin Nanocages: Study of Structure and Catalysis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7W7J
 
 | | Crystal structure of IrCp* immobilized apo-R52H-rHLFr (25 equiv) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Taher, M, Maity, B, Nakane, T, Abe, S, Mazumdar, S. | | Deposit date: | 2021-12-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Controlled Uptake of an Iridium Complex inside Engineered apo-Ferritin Nanocages: Study of Structure and Catalysis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7WYR
 
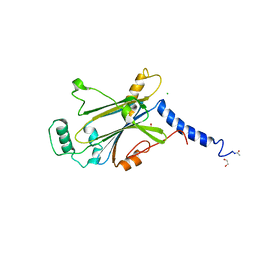 | | Crystal structure of Cypovirus Polyhedra mutant fused with CLN025 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin fused with CLN025 | | Authors: | Kojima, M, Abe, S, Hirata, K, Yamashita, K, Ueno, T. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering of an in-cell protein crystal for fastening a metastable conformation of a target miniprotein.
Biomater Sci, 11, 2023
|
|
7BON
 
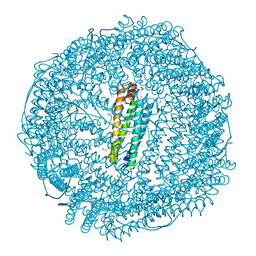 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7BOM
 
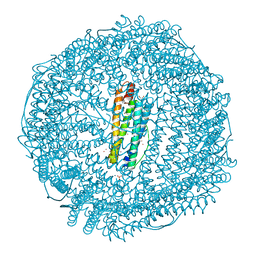 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr immobilized with gold ions. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7VIO
 
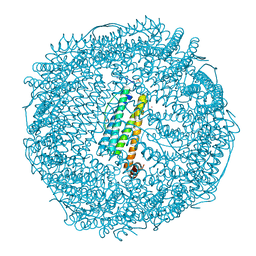 | | Crystal structure of recombinant horse spleen apo-R168H/L169C-Fr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Wu, J, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIU
 
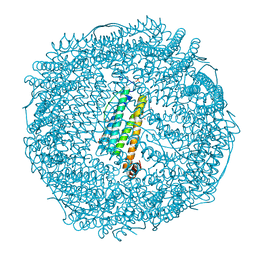 | | Crystal structure of Au(200EQ)-apo-R168C/L169C-rHLFr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIP
 
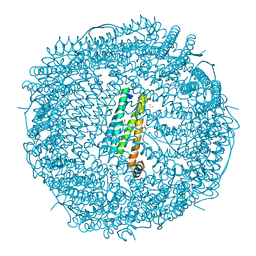 | | Crystal structure of Au(10EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIQ
 
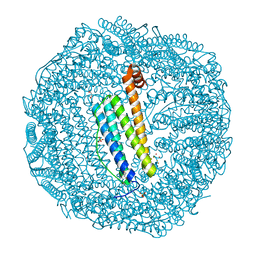 | | Crystal structure of Au(50EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIS
 
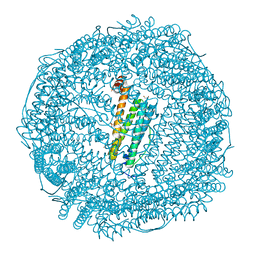 | | Crystal structure of Au(200EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIT
 
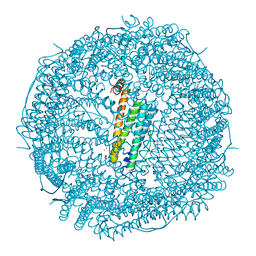 | | Crystal structure of Au(400EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
7VIR
 
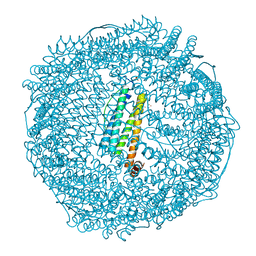 | | Crystal structure of Au(100EQ)-apo-R168H/L169C-rHLFr | | Descriptor: | CADMIUM ION, Ferritin light chain, GOLD ION, ... | | Authors: | Lu, C, Peng, X, Maity, B, Ito, N, Abe, S, Ueno, T, Lu, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of a gold clustering site in an engineered apo-ferritin cage.
Commun Chem, 5, 2022
|
|
2KUQ
 
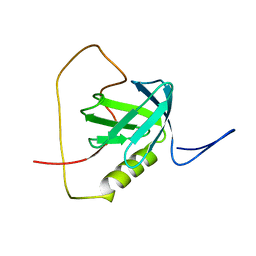 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | Fibroblast growth factor receptor substrate 3,LINKER,ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
7B81
 
 | |
8GST
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (pyruvate bound-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8GSR
 
 | | Crystal structure of L-2,4-diketo-3-deoxyrhamnonate hydrolase from Sphingomonas sp. (apo-form) | | Descriptor: | L-2,4-diketo-3-deoxyrhamnonate hydrolase, MAGNESIUM ION | | Authors: | Fukuhara, S, Watanabe, Y, Watanabe, S, Nishiwaki, H. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of l-2,4-Diketo-3-deoxyrhamnonate Hydrolase Involved in the Nonphosphorylated l-Rhamnose Pathway from Bacteria.
Biochemistry, 62, 2023
|
|
8Y11
 
 | |
8Y4J
 
 | |
8XWK
 
 | |
8Y4B
 
 | |
8Y46
 
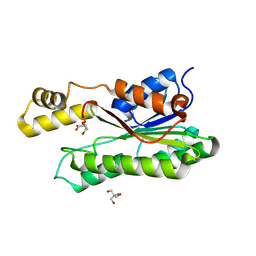 | | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase bound to L-KDF or L-2,4-DKDF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, L-2,4-diketo-3-deoxyfuconate, ... | | Authors: | Akagashi, M, Watanabe, S. | | Deposit date: | 2024-01-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structure of L-2-keto-3-deoxyfuconate 4-dehydrogenase reveals a unique binding mode as a alpha-furanosyl hemiketal of substrates.
Sci Rep, 14, 2024
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
2NZI
 
 | |
