2ZZW
 
 | | Crystal Structure of a Periplasmic Substrate Binding Protein in Complex with Zinc and Lactate | | Descriptor: | ABC transporter, solute-binding protein, LACTIC ACID, ... | | Authors: | Akiyama, N, Takeda, K, Miki, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a periplasmic substrate-binding protein in complex with calcium lactate
J.Mol.Biol., 392, 2009
|
|
3AAC
 
 | | Small heat shock protein hsp14.0 with the mutations of I120F and I122F in the form II crystal | | Descriptor: | Putative uncharacterized protein ST1653 | | Authors: | Takeda, K, Hayashi, T, Abe, T, Hirano, Y, Hanazono, Y, Yohda, M, Miki, K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dimer structure and conformational variability in the N-terminal region of an archaeal small heat shock protein, StHsp14.0
J.Struct.Biol., 174, 2011
|
|
3A5D
 
 | | Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase | | Descriptor: | V-type ATP synthase alpha chain, V-type ATP synthase beta chain, V-type ATP synthase subunit D, ... | | Authors: | Numoto, N, Hasegawa, Y, Takeda, K, Miki, K. | | Deposit date: | 2009-08-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Inter-subunit interaction and quaternary rearrangement defined by the central stalk of prokaryotic V1-ATPase
Embo Rep., 10, 2009
|
|
3AAB
 
 | | Small heat shock protein hsp14.0 with the mutations of I120F and I122F in the form I crystal | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Putative uncharacterized protein ST1653 | | Authors: | Takeda, K, Hayashi, T, Abe, T, Hirano, Y, Hanazono, Y, Yohda, M, Miki, K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Dimer structure and conformational variability in the N-terminal region of an archaeal small heat shock protein, StHsp14.0
J.Struct.Biol., 174, 2011
|
|
2ZHG
 
 | | Crystal structure of SoxR in complex with DNA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*DGP*DCP*DCP*DTP*DCP*DAP*DAP*DGP*DTP*DTP*DAP*DAP*DCP*DTP*DTP*DGP*DAP*DGP*DGP*DC)-3'), FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Watanabe, S, Kita, A, Kobayashi, K, Miki, K. | | Deposit date: | 2008-02-05 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3AZC
 
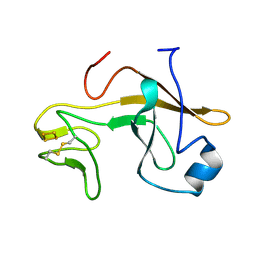 | | Crystal structure of the soluble part of cytochrome b6f complex iron-sulfur subunit from Thermosynechococcus elongatus BP-1 | | Descriptor: | Cytochrome b6-f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Veit, S, Takeda, K, Tsunoyama, Y, Roegner, M, Miki, K. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a thermophilic cyanobacterial b(6)f-type Rieske protein
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3A12
 
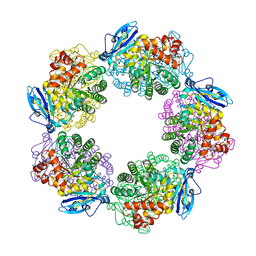 | | Crystal structure of Type III Rubisco complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
2ZDI
 
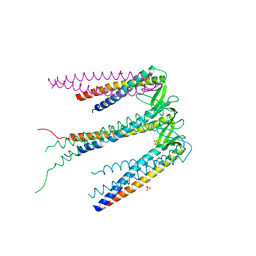 | | Crystal structure of Prefoldin from Pyrococcus horikoshii OT3 | | Descriptor: | Prefoldin subunit alpha, Prefoldin subunit beta, SULFATE ION | | Authors: | Kida, H, Miki, K. | | Deposit date: | 2007-11-23 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins
J.Mol.Biol., 376, 2008
|
|
3W5H
 
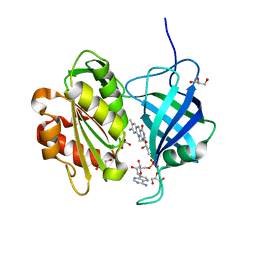 | | Ultra-high resolution structure of NADH-cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADH-cytochrome b5 reductase 3 | | Authors: | Takeda, K, Ohno, H, Kosugi, M, Takaba, K, Miki, K. | | Deposit date: | 2013-01-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2I
 
 | | Crystal structure of re-oxidized form (60 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2E
 
 | | Crystal structure of oxidation intermediate (20 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2G
 
 | | Crystal structure of fully reduced form of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2F
 
 | | Crystal structure of oxidation intermediate (10 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2H
 
 | | Crystal structure of oxidation intermediate (1min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3VQL
 
 | |
3W07
 
 | |
3W1Z
 
 | | Heat shock protein 16.0 from Schizosaccharomyces pombe | | Descriptor: | Heat shock protein 16 | | Authors: | Hanazono, Y, Takeda, K, Akiyama, N, Aikawa, Y, Miki, K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Nonequivalence Observed for the 16-Meric Structure of a Small Heat Shock Protein, SpHsp16.0, from Schizosaccharomyces pombe
Structure, 21, 2013
|
|
3VQK
 
 | |
3VX3
 
 | | Crystal structure of [NiFe] hydrogenase maturation protein HypB from Thermococcus kodakarensis KOD1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ... | | Authors: | Sasaki, D, Watanabe, S, Miki, K. | | Deposit date: | 2012-09-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure of a Novel Archaeal HypB for [NiFe] Hydrogenase Maturation
J.Mol.Biol., 425, 2013
|
|
3VQM
 
 | |
3WCU
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: Deoxygenated form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WCV
 
 | | The structure of a deoxygenated 400 kda hemoglobin provides a more accurate description of the cooperative mechanism of giant hemoglobins: CA bound form | | Descriptor: | A1 globin chain of giant V2 hemoglobin, A2 globin chain of giant V2 hemoglobin, B1 globin chain of giant V2 hemoglobin, ... | | Authors: | Numoto, N, Nakagawa, T, Ohara, R, Hasegawa, T, Kita, A, Yoshida, T, Maruyama, T, Imai, K, Fukumori, Y, Miki, K. | | Deposit date: | 2013-06-01 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a deoxygenated 400 kDa haemoglobin reveals ternary- and quaternary-structural changes of giant haemoglobins
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WHB
 
 | |
3WJU
 
 | |
3WJT
 
 | | Crystal structure of the L68D variant of mLolB | | Descriptor: | CHLORIDE ION, Outer-membrane lipoprotein LolB, SULFATE ION | | Authors: | Takeda, K, Tokuda, H, Miki, K. | | Deposit date: | 2013-10-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Roles of the Protruding Loop of Factor B Essential for the Localization of Lipoproteins (LolB) in the Anchoring of Bacterial Triacylated Proteins to the Outer Membran
J.Biol.Chem., 289, 2014
|
|
