2VVM
 
 | | The structure of MAO-N-D5, a variant of monoamine oxidase from Aspergillus niger. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N, ... | | Authors: | Atkin, K.E, Hart, S, Turkenburg, J.P, Brzozowski, A.M, Grogan, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Monoamine Oxidase from Aspergillus Niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution.
J.Mol.Biol., 384, 2008
|
|
2WBG
 
 | | Structure of family 1 beta-glucosidase from Thermotoga maritima in complex with 3-imino-2-oxa-(+)-castanospermine | | Descriptor: | (3Z,5S,6R,7S,8R,8aR)-3-(octylimino)hexahydro[1,3]oxazolo[3,4-a]pyridine-5,6,7,8-tetrol, ACETATE ION, BETA-GLUCOSIDASE A | | Authors: | Aguilar, M, Gloster, T.M, Turkenburg, J.P, Garcia-Moreno, M.I, Ortiz Mellet, C, Davies, G.J, Garcia Fernandez, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition by Ring-Modified Castanospermine Analogues: Tackling Enzyme Selectivity by Inhibitor Tailoring.
Org.Biomol.Chem., 7, 2009
|
|
2WH7
 
 | | The partial structure of a group A streptpcoccal phage-encoded tail fibre hyaluronate lyase Hylp2 | | Descriptor: | HYALURONIDASE-PHAGE ASSOCIATED | | Authors: | Martinez-Fleites, C, Black, G.W, Turkenburg, J.P, Smith, N.L, Taylor, E.J. | | Deposit date: | 2009-05-01 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Two Truncated Phage-Tail Hyaluronate Lyases from Streptococcus Pyogenes Serotype M1.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2X03
 
 | | The X-ray structure of the Streptomyces coelicolor A3 Chondroitin AC Lyase Y253A mutant | | Descriptor: | MAGNESIUM ION, PUTATIVE SECRETED LYASE | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-12-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
2WRU
 
 | | Semi-synthetic highly active analogue of human insulin NMeAlaB26-DTI- NH2 | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WS7
 
 | | Semi-synthetic analogue of human insulin ProB26-DTI | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Brzozowski, A.M, Jiracek, J, Zakova, L, Antolikova, E, Watson, C.J, Turkenburg, J.P, Dodson, G.G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Implications for the Active Form of Human Insulin Based on the Structural Convergence of Highly Active Hormone Analogues.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4TGL
 
 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
3ZHB
 
 | |
6ZPV
 
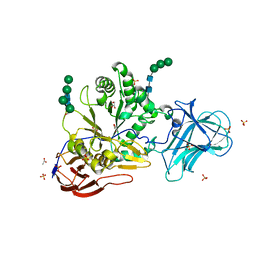 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZQ0
 
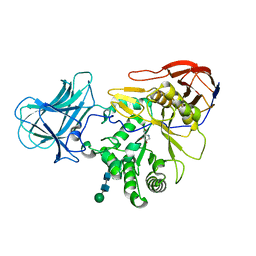 | | Structure of a-l-AraAZI-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPW
 
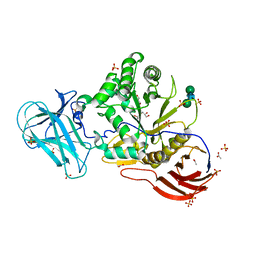 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPZ
 
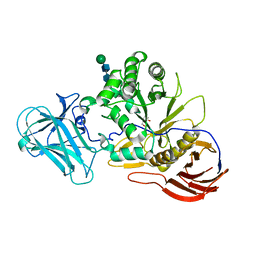 | | Structure of a-l-AraCS-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6Y5T
 
 | | Crystal structure of savinase at room temperature | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6Y5S
 
 | | Crystal structure of savinase at cryogenic conditions | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
6IAQ
 
 | | Structure of Amine Dehydrogenase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase N-terminus domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Grogan, G, Vaxelaire-Vergne, C, Beloti, L, Mayol, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6I3G
 
 | | Crystal structure of a putative peptide binding protein AppA from Clostridium difficile | | Descriptor: | ABC transporter, substrate-binding protein, family 5, ... | | Authors: | Hughes, A.M, Wilkinson, A, Dodson, E. | | Deposit date: | 2018-11-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the putative peptide-binding protein AppA from Clostridium difficile.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
258D
 
 | | FACTORS AFFECTING SEQUENCE SELECTIVITY ON NOGALAMYCIN INTERCALATION: THE CRYSTAL STRUCTURE OF D(TGTACA)-NOGALAMYCIN | | Descriptor: | ACETATE ION, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3'), NOGALAMYCIN, ... | | Authors: | Smith, C.K, Brannigan, J.A, Moore, M.H. | | Deposit date: | 1996-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Factors affecting DNA sequence selectivity of nogalamycin intercalation: the crystal structure of d(TGTACA)2-nogalamycin2.
J.Mol.Biol., 263, 1996
|
|
6H75
 
 | | SiaP A11N in complex with Neu5Ac (RT) | | Descriptor: | N-acetyl-beta-neuraminic acid, Sialic acid-binding periplasmic protein SiaP | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6H76
 
 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
3CTO
 
 | | Crystal Structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-04-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
3D55
 
 | | Crystal structure of M. tuberculosis YefM antitoxin | | Descriptor: | SULFATE ION, Uncharacterized protein Rv3357/MT3465 | | Authors: | Kumar, P, Issac, B, Dodson, E.J, Turkenberg, J.P, Mande, S.C. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein
J.Mol.Biol., 383, 2008
|
|
5LWH
 
 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
1H6X
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1H6Y
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
8ROZ
 
 | |
