3HRC
 
 | |
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3QGK
 
 | |
3QGA
 
 | |
1J6U
 
 | |
1J5S
 
 | |
4Q1G
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Polyketide biosynthesis enoyl-CoA isomerase PksI | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
4Q1H
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Polyketide biosynthesis enoyl-CoA isomerase PksI, ... | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
4Q1J
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, Polyketide biosynthesis enoyl-CoA isomerase PksI, SODIUM ION | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
1NF2
 
 | |
3DBS
 
 | | Structure of PI3K gamma in complex with GDC0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Wiesmann, C, Ultsch, M. | | Deposit date: | 2008-06-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer
J.Med.Chem., 51, 2008
|
|
1KQ3
 
 | | CRYSTAL STRUCTURE OF A GLYCEROL DEHYDROGENASE (TM0423) FROM THERMOTOGA MARITIMA AT 1.5 A RESOLUTION | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Wilson, I.A, Miller, M.D, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-01-03 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQ4
 
 | |
4Q1K
 
 | |
4Q1I
 
 | |
3CM9
 
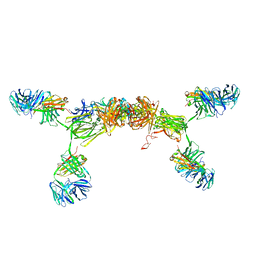 | | Solution Structure of Human SIgA2 | | Descriptor: | Immunoglobulin heavy chain, Immunoglobulin light chain, Secretory component | | Authors: | Bonner, A, Almogren, A, Furtado, P.B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2008-03-21 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The nonplanar secretory IgA2 and near planar secretory IgA1 solution structures rationalize their different mucosal immune responses.
J.Biol.Chem., 284, 2009
|
|
3LFF
 
 | | Human p38 MAP Kinase in Complex with RL166 | | Descriptor: | (4-{3-tert-butyl-5-[(1,3-thiazol-2-ylcarbamoyl)amino]-1H-pyrazol-1-yl}phenyl)acetic acid, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LFE
 
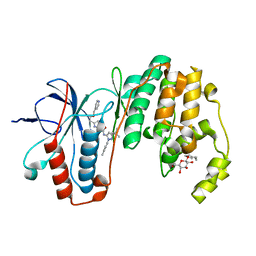 | | Human p38 MAP Kinase in Complex with RL116 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{4-[2-(pyridin-4-ylmethoxy)ethyl]-1,3-thiazol-2-yl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LFD
 
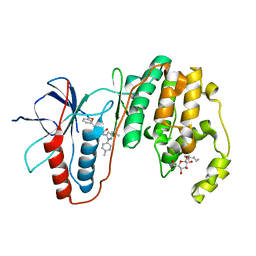 | | Human p38 MAP Kinase in Complex with RL113 | | Descriptor: | 1-{4-[2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LFC
 
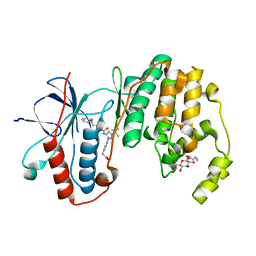 | | Human p38 MAP Kinase in Complex with RL99 | | Descriptor: | (4-{5-[({4-[2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}carbamoyl)amino]-3-tert-butyl-1H-pyrazol-1-yl}phenyl)acetic acid, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LFB
 
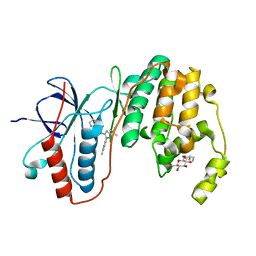 | | Human p38 MAP Kinase in Complex with RL98 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(1,3-thiazol-2-yl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3LFA
 
 | | Human p38 MAP Kinase in Complex with Dasatinib | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-01-16 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha
To be Published
|
|
3PG3
 
 | | Human p38 MAP Kinase in Complex with RL182 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{4-[2-(pyridin-3-ylmethoxy)ethyl]-1,3-thiazol-2-yl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of cell active N-pyrazole, N-thiazole urea inhibitors of the MAP kinase p38alpha
To be Published
|
|
2C69
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | (5Z)-5-(3-BROMOCYCLOHEXA-2,5-DIEN-1-YLIDENE)-N-(PYRIDIN-4-YLMETHYL)-1,5-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-AMINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: protein structure-guided design and SAR.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2C6O
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: protein structure-guided design and SAR.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
