5ZVX
 
 | |
5ZWA
 
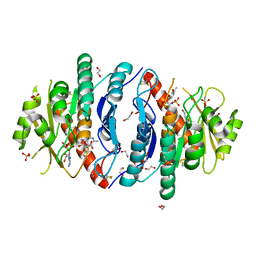 | | Crystal structure of Pyridoxal kinase (PdxK) from Salmonella typhimurium in complex with ADP, PL-linked to Lys233 via Schiff base in protomer A and the product (PLP) in protomer B | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Deka, G, Benazir, J.F, Kalyani, J.N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies on Salmonella typhimurium pyridoxal kinase: the first structural evidence for the formation of Schiff base with the substrate.
Febs J., 286, 2019
|
|
5ZVR
 
 | |
5ZUY
 
 | |
6AA9
 
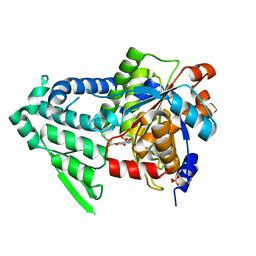 | | T166A mutant of D-Serine deaminase from Salmonella typhimurium | | Descriptor: | D-serine dehydratase, PHOSPHATE ION, SODIUM ION | | Authors: | Deka, G, Bharath, S.R, Shavithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-07-17 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on the decameric S. typhimurium arginine decarboxylase (ADC): Pyridoxal 5'-phosphate binding induces conformational changes
Biochem. Biophys. Res. Commun., 490, 2017
|
|
1UJQ
 
 | |
1VGA
 
 | | Structures of unligated and inhibitor complexes of W168F mutant of Triosephosphate Isomerase from Plasmodium falciparum | | Descriptor: | Triosephosphate isomerase | | Authors: | Eaazhisai, K, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2004-04-23 | | Release date: | 2004-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Unliganded and Inhibitor Complexes of W168F, a Loop6 Hinge Mutant of Plasmodium falciparum Triosephosphate Isomerase: Observation of an Intermediate Position of Loop6
J.Mol.Biol., 343, 2004
|
|
4D9E
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with L-cycloserine (LCS) | | Descriptor: | BENZAMIDINE, D-Cysteine desulfhydrase, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D96
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with 1-amino-1-carboxycyclopropane (ACC) | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9I
 
 | | Crystal structure of holo Diaminopropionate ammonia lyase from Escherichia coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diaminopropionate ammonia-lyase | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
4D92
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase soaked with beta-chloro-D-alanine shows pyruvate bound 4 A away from active site | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, PYRUVIC ACID, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9K
 
 | | Crystal structure of Escherichia coli Diaminopropionate ammonia lyase in apo form | | Descriptor: | Diaminopropionate ammonia-lyase, PHOSPHATE ION, SULFATE ION | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
4D99
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase with L-ser bound non-covalently at the active site | | Descriptor: | BENZAMIDINE, D-cysteine desulfhydrase, SERINE, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9N
 
 | | Crystal structure of Diaminopropionate ammonia lyase from Escherichia coli in complex with D-serine | | Descriptor: | D-SERINE, Diaminopropionate ammonia-lyase | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
4D9B
 
 | | Pyridoxamine 5' phosphate (PMP) bound form of Salmonella typhimurium D-Cysteine desulfhydrase obtained after co-crystallization with D-cycloserine | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9C
 
 | | PMP bound form of Salmonella typhimurium D-Cysteine desulfhydrase obtained after co-crystallization with L-cycloserine | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, D-Cysteine desulfhydrase | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D8T
 
 | | Crystal structure of D-Cysteine desulfhydrase from Salmonella typhimurium at 2.2 A resolution | | Descriptor: | BENZAMIDINE, CHLORIDE ION, D-cysteine desulfhydrase, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9F
 
 | | D-Cysteine desulfhydrase from Salmonella typhimurium complexed with D-cycloserine (DCS) | | Descriptor: | BENZAMIDINE, D-Cysteine desulfhydrase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D8U
 
 | | Crystal structure of D-Cysteine desulfhydrase from Salmonella typhimurium at 3.3 A in monoclinic space group with 8 subunits in the asymmetric unit | | Descriptor: | D-cysteine desulfhydrase, PHOSPHATE ION | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9M
 
 | | Crystal structure of Diaminopropionate ammonia lyase from Escherichia coli in complex with aminoacrylate-PLP azomethine reaction intermediate | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, Diaminopropionate ammonia-lyase | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
4D97
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase with D-ser bound at active site | | Descriptor: | BENZAMIDINE, D-SERINE, D-cysteine desulfhydrase | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D8W
 
 | | Salmonella typhimurium D-Cysteine desulfhydrase soaked with D-cys shows pyruvate bound 4 A away from active site | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Bharath, S.R, Shveta, B, Rajesh, K.H, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Mutational Studies on Substrate Specificity and Catalysis of Salmonella typhimurium D-Cysteine Desulfhydrase.
Plos One, 7, 2012
|
|
4D9G
 
 | | Crystal structure of Selenomethionine incorporated holo Diaminopropionate ammonia lyase from Escherichia coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative diaminopropionate ammonia-lyase | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
4EGV
 
 | |
4GPD
 
 | |
