1OYU
 
 | |
1PE8
 
 | | Thermolysin with monocyclic inhibitor | | Descriptor: | 2-ETHOXYETHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | Authors: | Juers, D, Pyun, H.-J, Bartlett, P.A, Matthews, B.W. | | Deposit date: | 2003-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
1OV7
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 2-Allyl-6-Methyl-Phenol | | Descriptor: | 2-ALLYL-6-METHYL-PHENOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1OVJ
 
 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound with 3-Fluoro-2-Methyl_Aniline | | Descriptor: | 3-FLUORO-2-METHYL-ANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-26 | | Release date: | 2004-04-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1PE5
 
 | | Thermolysin with tricyclic inhibitor | | Descriptor: | (6-METHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL)METHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | Authors: | Juers, D, Holland, D, Morgan, B.P, Bartlett, P.A, Matthews, B.W. | | Deposit date: | 2003-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
1PE7
 
 | | Thermolysin with bicyclic inhibitor | | Descriptor: | 2-(4-METHYLPHENOXY)ETHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | Authors: | Juers, D, Yusuff, N, Bartlett, P.A, Matthews, B.W. | | Deposit date: | 2003-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
1TLA
 
 | | HYDROPHOBIC CORE REPACKING AND AROMATIC-AROMATIC INTERACTION IN THE THERMOSTABLE MUTANT OF T4 LYSOZYME SER 117 (RIGHT ARROW) PHE | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, T4 LYSOZYME | | Authors: | Anderson, D.E, Hurley, J.H, Nicholson, H, Baase, W.A, Matthews, B.W. | | Deposit date: | 1993-03-22 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophobic core repacking and aromatic-aromatic interaction in the thermostable mutant of T4 lysozyme Ser 117-->Phe.
Protein Sci., 2, 1993
|
|
1ZDP
 
 | | Crystal Structure Analysis of Thermolysin Complexed with the Inhibitor (S)-thiorphan | | Descriptor: | (2-MERCAPTOMETHYL-3-PHENYL-PROPIONYL)-GLYCINE, CALCIUM ION, Thermolysin, ... | | Authors: | Roderick, S.L, Fournie-Zaluski, M.C, Roques, B.P, Matthews, B.W. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiorphan and retro-thiorphan display equivalent interactions when bound to crystalline thermolysin
Biochemistry, 28, 1989
|
|
1Z9G
 
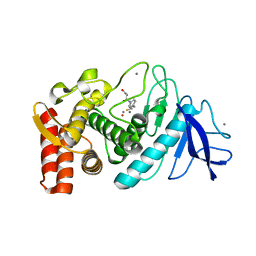 | | Crystal Structure Analysis of Thermolysin Complexed with the Inhibitor (R)-retro-thiorphan | | Descriptor: | (R)-RETRO-THIORPHAN, CALCIUM ION, Thermolysin, ... | | Authors: | Roderick, S.L, Fournie-Zaluski, M.C, Roques, B.P, Matthews, B.W. | | Deposit date: | 2005-04-01 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thiorphan and retro-thiorphan display equivalent interactions when bound to crystalline thermolysin
Biochemistry, 28, 1989
|
|
2B73
 
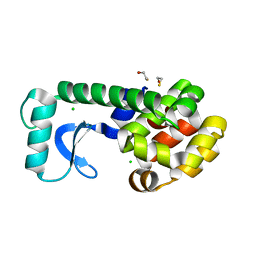 | | T4 Lysozyme mutant L99A at 100 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B3L
 
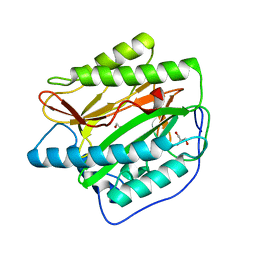 | | Crystal structure of type I human methionine aminopeptidase in the apo form | | Descriptor: | ACETIC ACID, GLYCEROL, Methionine aminopeptidase 1, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B3K
 
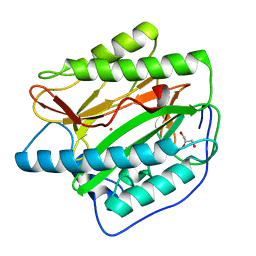 | | Crystal structure of Human Methionine Aminopeptidase Type I in the holo form | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B70
 
 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B75
 
 | | T4 Lysozyme mutant L99A at 150 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6Z
 
 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B3H
 
 | | Crystal structure of Human Methionine Aminopeptidase Type I with a third cobalt in the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Addlagatta, A, Hu, X, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis for the Functional Differences between Type I and Type II Human Methionine Aminopeptidases(,).
Biochemistry, 44, 2005
|
|
2B6W
 
 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B72
 
 | | T4 Lysozyme mutant L99A at 100 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6X
 
 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6Y
 
 | | T4 Lysozyme mutant L99A at ambient pressure | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2B6T
 
 | | T4 Lysozyme mutant L99A at 200 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural rigidity of a large cavity-containing protein revealed by high-pressure crystallography.
J.Mol.Biol., 367, 2007
|
|
2B74
 
 | | T4 Lysozyme mutant L99A at 100 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | Deposit date: | 2005-10-03 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1HEL
 
 | |
1HEM
 
 | |
1HER
 
 | |
