4MK3
 
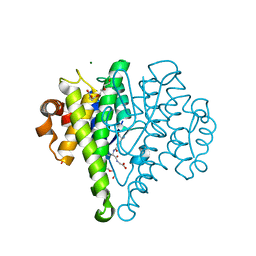 | | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h) | | Descriptor: | Glutathione S-transferase, L-GAMMA-GLUTAMYL-3-SULFINO-L-ALANYLGLYCINE, MAGNESIUM ION, ... | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Cupriavidus metallidurans CH34, target EFI-507362, with bound glutathione sulfinic acid (gso2h)
To be published
|
|
4MP4
 
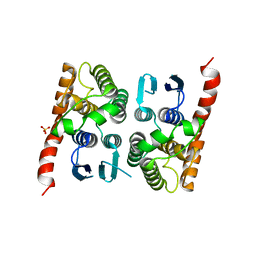 | | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Acinetobacter baumannii, Target EFI-501785, apo structure
To be Published
|
|
4F4R
 
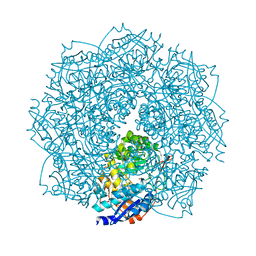 | | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of D-mannonate dehydratase homolog from Chromohalobacter salexigens (Target EFI-502114), with bound NA, ordered loop
to be published
|
|
4DYE
 
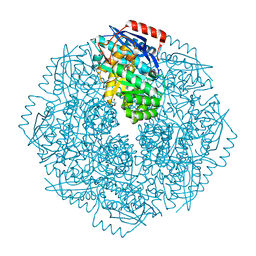 | | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, isomerase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop
to be published
|
|
4IMR
 
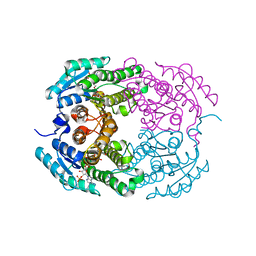 | | Crystal structure of 3-oxoacyl (acyl-carrier-protein) reductase (target EFI-506442) from agrobacterium tumefaciens C58 with NADP bound | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of 3-Oxoacyl (Acyl-Carrier-Protein) Reductase Atu5465 from Agrobacterium Tumefaciens
To be Published
|
|
4JXK
 
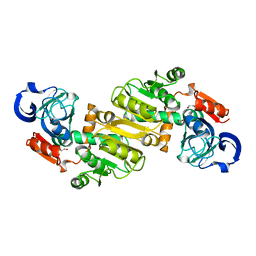 | | Crystal Structure of Oxidoreductase ROP_24000 (Target EFI-506400) from Rhodococcus opacus B4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, oxidoreductase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Oxidoreductase Rop_24000 (Target EFI-506400) from Rhodococcus Opacus
To be Published
|
|
4JXY
 
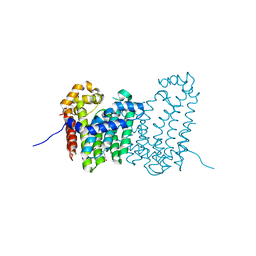 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE PATL_3739 (TARGET EFI-509195) FROM Pseudoalteromonas atlantica | | Descriptor: | GLUTARIC ACID, PHOSPHATE ION, Trans-hexaprenyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF ISOPRENOID SYNTHASE PATL_3739 FROM Pseudoalteromonas atlantica
To be Published
|
|
4JBB
 
 | | Crystal structure of Glutathione S-transferase A6TBY7(Target EFI-507184) from Klebsiella pneumoniae MGH 78578, GSH complex | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase A6Tby7 (Target Efi-507184) from Klebsiella Pneumoniae
To be Published
|
|
4JED
 
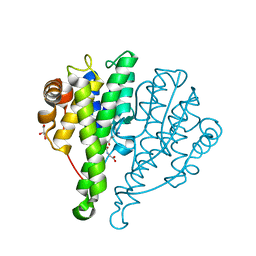 | | Crystal structure of glutathione s-transferase mrad2831_1084 (target efi-507060) from methylobacterium radiotolerans jcm 2831, complex with glutathione sulfonate | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of glutathione S-transferase (TARGET EFI-507060) from Methylobacterium radiotolerans
To be Published
|
|
4DO7
 
 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2 | | Descriptor: | Amidohydrolase 2, SULFATE ION, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2
to be published
|
|
4JYX
 
 | | Crystal structure of polyprenyl synthase PATL_3739 (TARGET EFI-509195) FROM PSEUDOALTEROMONAS ATLANTICA, COMPLEX WITH INORGANIC PHOSPHATE AND AN UNKNOWN LIGAND | | Descriptor: | PHOSPHATE ION, Trans-hexaprenyltranstransferase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-01 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Patl_3739 from Pseudoalteromonas Atlantica
To be Published
|
|
4IQ1
 
 | | Crystal structure of glutathione s-transferase MHA_0454 (TARGET EFI-507015) FROM Mannheimia haemolytica, SUBSTRATE-FREE | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, AL Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of probable glutathione s-transferase MHA_0454 (TARGET EFI-507015) FROM Mannheimia haemolytica, Substrate-free
To be Published
|
|
4IW9
 
 | | Crystal structure of glutathione s-transferase mha_0454 (target efi-507015) from mannheimia haemolytica, gsh complex | | Descriptor: | ACETATE ION, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Probable Glutathione S-Transferase Mha_0454 (Target Efi-507015) from Mannheimia Haemolytica
To be Published
|
|
4GGH
 
 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221)
To be Published
|
|
4JDP
 
 | | Crystal structure of probable p-nitrophenyl phosphatase (pho2) (target EFI-501307) from Archaeoglobus fulgidus DSM 4304 with Magnesium bound | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Allen, K.N, Dunaway-Mariano, D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of probable p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
4NHZ
 
 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290, with one glutathione bound | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structure of Glutathione Transferase Bbta-3750 from Bradyrhizobium Sp., Target Efi-507290
To be Published
|
|
4G9H
 
 | | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target EFI-501894, with bound glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target efi-501894, with bound glutathione
To be Published
|
|
4IHC
 
 | | Crystal structure of probable mannonate dehydratase Dd703_0947 (target EFI-502222) from Dickeya dadantii Ech703 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mannonate dehydratase Dd703_0947 from Dickeya dadantii Ech703
To be Published
|
|
4G9B
 
 | | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid | | Descriptor: | Beta-phosphoglucomutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
To be Published
|
|
4INF
 
 | | Crystal structure of amidohydrolase saro_0799 (target efi-505250) from novosphingobium aromaticivorans dsm 12444 with bound calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of amidohydrolase sarp_0799 (target efi-505250) from novosphingobium aromaticivorans
To be Published
|
|
4GIR
 
 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, ethylene glycol and sulfate bound (ordered loops, space group P41212) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enolase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, ethylene glycol and sulfate bound (ordered loops, space group P41212)
To be Published
|
|
4JXN
 
 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE ISP_B (TARGET EFI-509198) FROM Roseobacter denitrificans | | Descriptor: | CHLORIDE ION, Octaprenyl-diphosphate synthase, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Isoprenoid Synthase Isp_B from Roseobacter Denitrificans
To be Published
|
|
4EBU
 
 | | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-24 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502312) from Oceanicola granulosus, with bound AMP/ADP crystal form I
TO BE PUBLISHED
|
|
4O7H
 
 | | Crystal structure of a glutathione S-transferase from Rhodospirillum rubrum F11, Target EFI-507460 | | Descriptor: | Glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Attonito, J.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-24 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione S-transferase from Rhodospirillum rubrum F11, Target EFI-507460
TO BE PUBLISHED
|
|
4EUM
 
 | | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II
To be Published
|
|
