8S2V
 
 | |
8S2U
 
 | |
8S2W
 
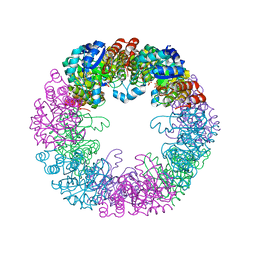 | |
9MRW
 
 | | Functional Implications of Hexameric Dynamics in SARS-CoV-2 Nsp15 | | Descriptor: | Uridylate-specific endoribonuclease nsp15 | | Authors: | Ketawala, G.K, Sonowal, M, Schrag, L, Fromme, R, Botha, S, Fromme, P. | | Deposit date: | 2025-01-08 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional implications of hexameric dynamics in SARS-CoV-2 Nsp15.
Protein Sci., 34, 2025
|
|
9MRY
 
 | | Functional Implications of HexamericDynamics in SARS-CoV-2 Nsp15 | | Descriptor: | Uridylate-specific endoribonuclease nsp15 | | Authors: | Ketawala, G.K, Sonowal, M, Schrag, L, Fromme, R, Botha, S, Fromme, P. | | Deposit date: | 2025-01-09 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional implications of hexameric dynamics in SARS-CoV-2 Nsp15.
Protein Sci., 34, 2025
|
|
5BUP
 
 | | Crystal structure of the ZP-C domain of mouse ZP2 | | Descriptor: | ACETATE ION, Zona pellucida sperm-binding protein 2 | | Authors: | Nishimura, K, Jovine, L. | | Deposit date: | 2015-06-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9FA5
 
 | | Lysozyme structure at room temperature by serial synchrotron crystallography with COC chips | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Lysozyme C, ... | | Authors: | Quereda-Moraleda, I, Grieco, A, Botha, S, Manna, A, Ros, A, Martin-Garcia, J.M. | | Deposit date: | 2024-05-10 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cyclic Olefin Copolymer-Based Fixed-Target Sample Delivery Device for Protein X-ray Crystallography.
Anal.Chem., 96, 2024
|
|
4BOH
 
 | | Madanins (MEROPS I53) are cleaved by thrombin and factor Xa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, SULFATE ION, ... | | Authors: | Figueiredo, A.C, deSanctis, D, Pereira, P.J.B. | | Deposit date: | 2013-05-20 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The Tick-Derived Anticoagulant Madanin is Processed by Thrombin and Factor Xa.
Plos One, 8, 2013
|
|
9NCC
 
 | |
9ORS
 
 | |
9OR7
 
 | | X-ray diffraction structure of CTX-M-14 beta-lactamase soaked with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, PHOSPHATE ION | | Authors: | Vlahakis, N.W, Rodriguez, J.A, Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2025-05-21 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining MicroED and native mass spectrometry for structural discovery of enzyme-small molecule complexes
To Be Published
|
|
9OR3
 
 | | X-ray diffraction structure of CTX-M-14 beta-lactamase co-crystallized with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, GLYCEROL, ... | | Authors: | Vlahakis, N.W, Rodriguez, J.A, Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2025-05-21 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining MicroED and native mass spectrometry for structural discovery of enzyme-small molecule complexes
To Be Published
|
|
9ORG
 
 | |
9ORY
 
 | |
9NAT
 
 | |
9ORB
 
 | | X-ray diffraction structure of the CTX-M-14 beta-lactamase-avibactam complex an inhibitor cocktail-soaked crystal | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, PHOSPHATE ION | | Authors: | Vlahakis, N.W, Rodriguez, J.A, Jacobs, L.M.C, Chen, Y. | | Deposit date: | 2025-05-21 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining MicroED and native mass spectrometry for structural discovery of enzyme-small molecule complexes
To Be Published
|
|
9ORH
 
 | |
9OQE
 
 | |
9ORL
 
 | |
9ORV
 
 | | X-ray diffraction structure of lysozyme co-crystallized with N,N',N"-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Flowers, C.W, Vlahakis, N.W, Rodriguez, J.A. | | Deposit date: | 2025-05-22 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining MicroED and native mass spectrometry for structural discovery of enzyme-small molecule complexes
To Be Published
|
|
9ORX
 
 | |
9ORZ
 
 | |
9OS0
 
 | |
9OS1
 
 | |
9OS8
 
 | |
