4JFM
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate | | Descriptor: | 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFJ
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFK
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4M8M
 
 | |
4FAF
 
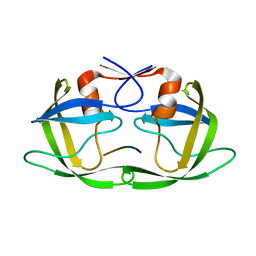 | | Substrate CA/p2 in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, substrate CA/p2 peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
5Y7L
 
 | |
4FAE
 
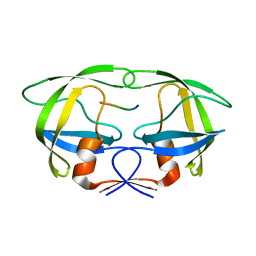 | | Substrate p2/NC in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, Substrate p2/NC peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
8XIQ
 
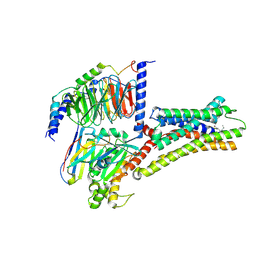 | | Structure of L796778-SSTR3 G protein complex | | Descriptor: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
8XIO
 
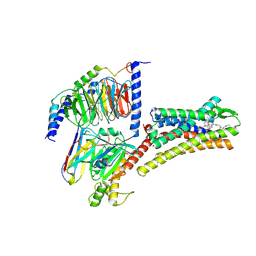 | | Structure of L797591-SSTR1 G protein complex | | Descriptor: | (2~{S})-~{N}-[(4~{S})-6-azanyl-2,2,4-trimethyl-hexyl]-3-naphthalen-1-yl-2-[[2-phenylethyl(2-pyridin-2-ylethyl)carbamoyl]amino]propanamide, G-alpha-i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
8XIR
 
 | | Structure of pasireotide-SSTR3 G protein complex | | Descriptor: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
8XIP
 
 | | Structure of Pasireotide-SSTR1 G protein complex | | Descriptor: | 004-DTR-LYS-TY5-PHE-A1D5E, G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
7WOH
 
 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(2S)-3-phenyl-2-[[(E)-3-phenylprop-2-enoyl]amino]propanoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOF
 
 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO1
 
 | | Discovery of SARS-CoV-2 3CLpro peptidomimetic inhibitors through H41-specific protein-ligand interactions | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]cyclohexanecarboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO3
 
 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-2-[[(2S)-2-[[(E)-3-(4-methoxyphenyl)prop-2-enoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO2
 
 | | SARS-CoV-2 3CLPro Peptidomimetic Inhibitor TPM5 | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S}-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]furan-2-carboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
5GZX
 
 | |
5H00
 
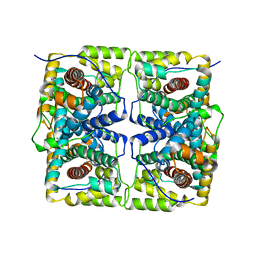 | |
5GZY
 
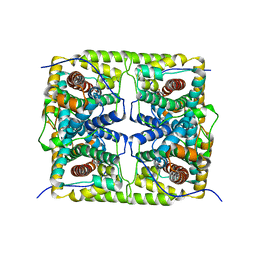 | |
5H6Y
 
 | |
8HK5
 
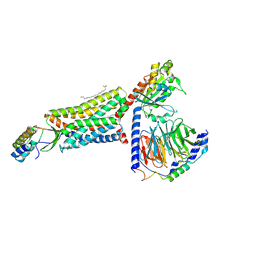 | | C5aR1-Gi-C5a protein complex | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, Complement C5, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK3
 
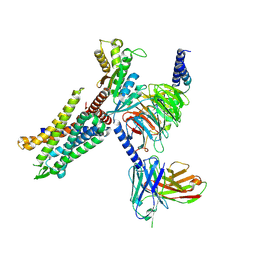 | | C3aR-Gi-apo protein complex | | Descriptor: | C3a anaphylatoxin chemotactic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
8HK2
 
 | | C3aR-Gi-C3a protein complex | | Descriptor: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | Authors: | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
7K6B
 
 | |
7KRB
 
 | |
