4CXY
 
 | | Crystal structure of human FTO in complex with acylhydrazine inhibitor 21 | | Descriptor: | (E)-4-(2-Nicotinoylhydrazinyl)-4-oxobut-2-enoic acid, ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE FTO, NICKEL (II) ION | | Authors: | Toh, D.W, Sun, L, Tan, J, Chen, Y, Lau, L.Z.M, Hong, W, Woon, E.C.Y, Gao, Y.G. | | Deposit date: | 2014-04-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor ofN6-methyladenosine demethylase FTO.
Chem Sci, 6, 2015
|
|
4CXX
 
 | | Crystal structure of human FTO in complex with acylhydrazine inhibitor 16 | | Descriptor: | (2E)-4-{N'-[4-(4-tert-Butyl-benzyl)-pyridine-3-carbonyl]-hydrazino}-4-oxo-but-2-enoic acid, ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE FTO, NICKEL (II) ION | | Authors: | Toh, D.W, Sun, L, Tan, J, Chen, Y, Lau, L.Z.M, Hong, W, Woon, E.C.Y, Gao, Y.G. | | Deposit date: | 2014-04-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor ofN6-methyladenosine demethylase FTO.
Chem Sci, 6, 2015
|
|
4CXW
 
 | | Crystal structure of human FTO in complex with subfamily-selective inhibitor 12 | | Descriptor: | (2E)-4-[N'-(4-benzyl-pyridine-3-carbonyl)-hydrazino]-4-oxo-but-2-enoic acid, ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE FTO, NICKEL (II) ION | | Authors: | Toh, D.W, Sun, L, Tan, J, Chen, Y, Lau, L.Z.M, Hong, W, Woon, E.C.Y, Gao, Y.G. | | Deposit date: | 2014-04-09 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor ofN6-methyladenosine demethylase FTO.
Chem Sci, 6, 2015
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
6F2S
 
 | | CryoEM structure of Ageratum Yellow Vein virus (AYVV) | | Descriptor: | Capsid protein, coat protein subunit H, coat protein subunit I, ... | | Authors: | Hesketh, E.L, Saunders, K, Fisher, C, Potze, J, Stanley, J, Lomonossoff, G.P, Ranson, N.A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The 3.3 angstrom structure of a plant geminivirus using cryo-EM.
Nat Commun, 9, 2018
|
|
5JXV
 
 | | Solid-state MAS NMR structure of immunoglobulin beta 1 binding domain of protein G (GB1) | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Andreas, L.B, Jaudzems, K, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JZR
 
 | | Solid-state MAS NMR structure of Acinetobacter phage 205 (AP205) coat protein in assembled capsid particles | | Descriptor: | Coat protein | | Authors: | Jaudzems, K, Andreas, L.B, Stanek, J, Lalli, D, Bertarello, A, Le Marchand, T, Cala-De Paepe, D, Kotelovica, S, Akopjana, I, Knott, B, Wegner, S, Engelke, F, Lesage, A, Emsley, L, Tars, K, Herrmann, T, Pintacuda, G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure of fully protonated proteins by proton-detected magic-angle spinning NMR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8ZCI
 
 | | X-ray structure of the human heart fatty acid-binding protein complexed with R-Ibuprofen | | Descriptor: | (2R)-2-[4-(2-methylpropyl)phenyl]propanoic acid, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sugiyama, S, Tanaka, J, Matsuoka, S, Murata, M. | | Deposit date: | 2024-04-29 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with R-Ibuprofen
To Be Published
|
|
6TK2
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 1ms structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK1
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 20ms structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK5
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 800fs+2ps structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK7
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: Dark structure in acidic conditions | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6T68
 
 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | CHLORIDE ION, MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
6T4R
 
 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
2VKM
 
 | | Crystal structure of GRL-8234 bound to BACE (Beta-secretase) | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Hong, L, Tang, J, Ghosh, A.K. | | Deposit date: | 2007-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent Memapsin 2 (Beta-Secretase) Inhibitors: Design, Synthesis, Protein-Ligand X-Ray Structure, and in Vivo Evaluation.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2HYY
 
 | | Human Abl kinase domain in complex with imatinib (STI571, Glivec) | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2HZ4
 
 | | Abl kinase domain unligated and in complex with tetrahydrostaurosporine | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Liebetanz, J, Fabbro, D, Manley, P. | | Deposit date: | 2006-08-08 | | Release date: | 2007-01-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia.
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
4CKU
 
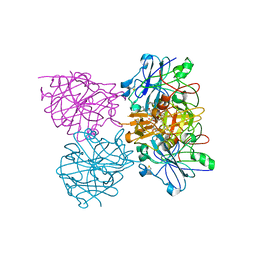 | | Three dimensional structure of plasmepsin II in complex with hydroxyethylamine-based inhibitor | | Descriptor: | 5-[1,1-bis(oxidanylidene)-1,2-thiazinan-2-yl]-N3-[(2S,3R)-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-butan-2-yl]-N1,N1-dipropyl-benzene-1,3-dicarboxamide, PLASMEPSIN-2 | | Authors: | Tars, K, Leitans, J, Jaudzems, K. | | Deposit date: | 2014-01-08 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasmepsin Inhibitory Activity and Structure-Guided Optimization of a Potent Hydroxyethylamine-Based Antimalarial Hit.
Acs Med.Chem.Lett., 5, 2014
|
|
2M2Q
 
 | | Solution structure of MCh-1: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-1 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
2M2R
 
 | | Solution structure of MCh-2: A novel inhibitor cystine knot peptide from Momordica charantia | | Descriptor: | Inhibitor cystine knot peptide MCh-2 | | Authors: | He, W, Chan, L, Clark, R.J, Tang, J, Zeng, G, Franco, O.L, Cantacessi, C, Craik, D.J, Daly, N.L, Tan, N. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Novel Inhibitor Cystine Knot Peptides from Momordica charantia.
Plos One, 8, 2013
|
|
9FSE
 
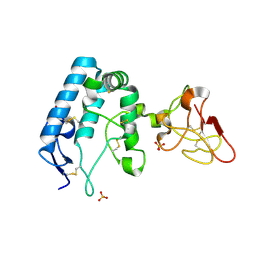 | | Human ROR2 cysteine-rich domain (CRD) and Kringle domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase transmembrane receptor ROR2 | | Authors: | Griffiths, S.C, Tan, J, Wagner, A, Blazer, L.L, Adams, J.J, Srinivasan, S, Moghisaei, S, Sidhu, S.S, Siebold, C, Ho, H.H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and function of the ROR2 cysteine-rich domain in vertebrate noncanonical WNT5A signaling.
Elife, 13, 2024
|
|
7Y7Y
 
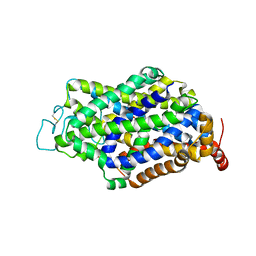 | | Cryo-EM structure of human GABA transporter GAT1 bound with nipecotic acid in NaCl solution in an inward-occluded state at 2.4 angstrom | | Descriptor: | (3R)-piperidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7W
 
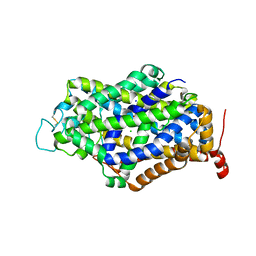 | | Cryo-EM structure of human GABA transporter GAT1 bound with GABA in NaCl solution in an inward-occluded state at 2.4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7V
 
 | | Cryo-EM structure of human apo GABA transporter GAT1 in an inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Sodium- and chloride-dependent GABA transporter 1 | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
