2B5R
 
 | | 1B Lactamase / B Lactamase Inhibitor | | Descriptor: | Beta-lactamase TEM, Beta-lactamase inhibitory protein | | Authors: | Rahat, O, Albeck, S, Meged, R, Dym, O, Screiber, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-09-29 | | Release date: | 2006-04-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding Hot Spots in the TEM1-BLIP Interface in Light of its Modular Architecture.
J.Mol.Biol., 102, 2006
|
|
1S0W
 
 | |
7QU5
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QTY
 
 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | Descriptor: | 1-(furan-2-ylmethyl)-3-(2-methylphenyl)thiourea, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QU3
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QU0
 
 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8OVY
 
 | | Structure of analogue of superfolded GFP | | Descriptor: | Green fluorescent protein | | Authors: | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
8CMY
 
 | |
7MEQ
 
 | | Crystal structure of human TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Transmembrane protease serine 2, ... | | Authors: | Fraser, B, Beldar, S, Hutchinson, A, Li, Y, Seitova, A, Edwards, A.M, Benard, F, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Nat.Chem.Biol., 18, 2022
|
|
7Q22
 
 | | cryo iDPC-STEM structure recorded with CSA 2.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2Q
 
 | | cryo iDPC-STEM structure recorded with CSA 3.5 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2S
 
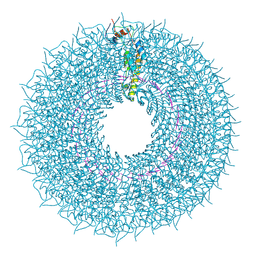 | | cryo iDPC-STEM structure recorded with CSA 4.5 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q23
 
 | | cryo iDPC-STEM structure recorded with CSA 3.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
7Q2R
 
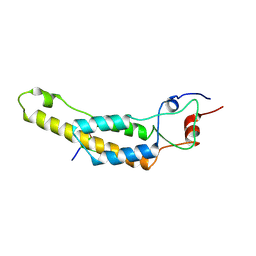 | | cryo iDPC-STEM structure recorded with CSA 4.0 | | Descriptor: | Capsid protein, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Sachse, C, Leidl, M.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single-particle cryo-EM structures from iDPC-STEM at near-atomic resolution.
Nat.Methods, 19, 2022
|
|
6Z4R
 
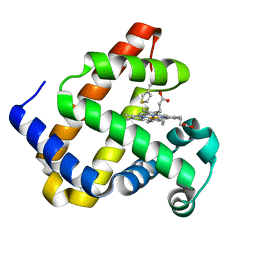 | |
6Z4T
 
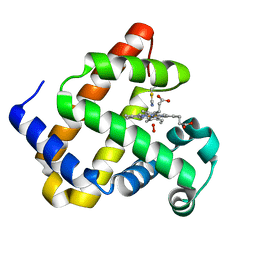 | |
2Q5A
 
 | | human Pin1 bound to L-PEPTIDE | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
1QL9
 
 | |
7NPD
 
 | | Vibiro cholerae ParA2 | | Descriptor: | Walker A-type ATPase | | Authors: | Parker, A.V, Bergeron, J.R.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the bacterial DNA segregation ATPase filament reveals the conformational plasticity of ParA upon DNA binding.
Nat Commun, 12, 2021
|
|
6Z8R
 
 | | Copper transporter OprC | | Descriptor: | COPPER (II) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8U
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8S
 
 | | Copper transporter OprC | | Descriptor: | COPPER (I) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8Q
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8Z
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z8T
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
