2MEA
 
 | |
8JL8
 
 | | Crystal structure of the collagen binding domain of Cnm from Streptococcus mutans | | Descriptor: | Collagen-binding adhesin, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S.-i, Hirata, A, Takano, K. | | Deposit date: | 2023-06-02 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure, Stability and Binding Properties of Collagen-Binding Domains from Streptococcus mutans.
Chemistry, 5, 2023
|
|
5X7K
 
 | |
3U3G
 
 | | Structure of LC11-RNase H1 Isolated from Compost by Metagenomic Approach: Insight into the Structural Bases for Unusual Enzymatic Properties of Sto-RNase H1 | | Descriptor: | CHLORIDE ION, Ribonuclease H, UNKNOWN LIGAND | | Authors: | Nguyen, T.N, Angkawidjaja, C, Kanaya, E, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activity, stability, and structure of metagenome-derived LC11-RNase H1, a homolog of Sulfolobus tokodaii RNase H1
Protein Sci., 21, 2012
|
|
2YV0
 
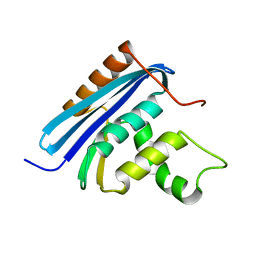 | | Structural and Thermodynamic Analyses of E. coli ribonuclease HI Variant with Quintuple Thermostabilizing Mutations | | Descriptor: | Ribonuclease HI | | Authors: | Haruki, M, Motegi, T, Tadokoro, T, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-04-06 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and thermodynamic analyses of Escherichia coli RNase HI variant with quintuple thermostabilizing mutations.
Febs J., 274, 2007
|
|
4OEC
 
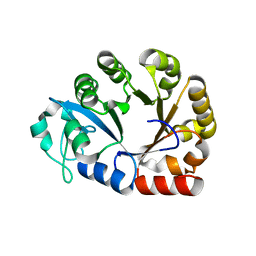 | | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1 | | Descriptor: | Glycerophosphoryl diester phosphodiesterase, MAGNESIUM ION | | Authors: | Atsuta, Y, You, D.J, Takano, K, Koga, Y, Kanaya, S. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glycerophosphodiester phosphodiesterase from Thermococcus kodakarensis KOD1
To be Published
|
|
6KTK
 
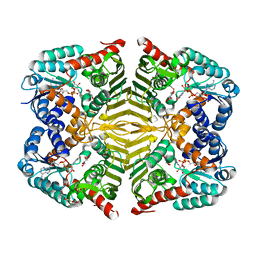 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTL
 
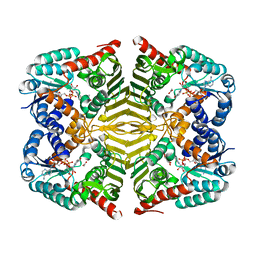 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTJ
 
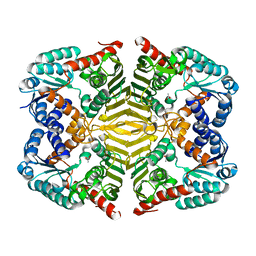 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, apo-form, from Paracoccus laeviglucosivorans | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5AVH
 
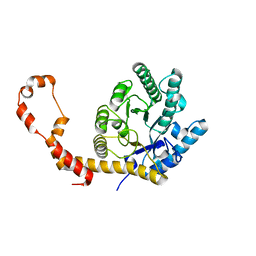 | | The 0.90 angstrom structure (I222) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | Xylose isomerase | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVN
 
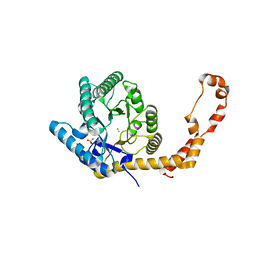 | | The 1.03 angstrom structure (P212121) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVD
 
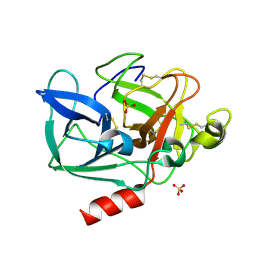 | | The 0.86 angstrom structure of elastase crystallized in high-strength agarose hydrogel | | Descriptor: | Chymotrypsin-like elastase family member 1, SULFATE ION | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5AVG
 
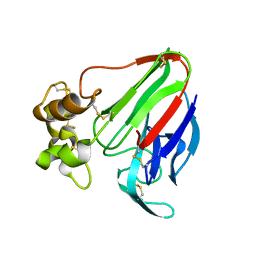 | | The 0.95 angstrom structure of thaumatin crystallized in high-strength agarose hydrogel | | Descriptor: | Thaumatin-1 | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, M, Sazaki, G, Hirose, M, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
8WHC
 
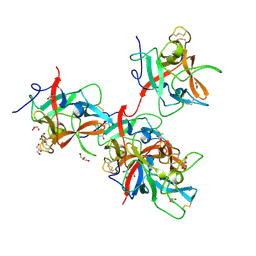 | |
8WIO
 
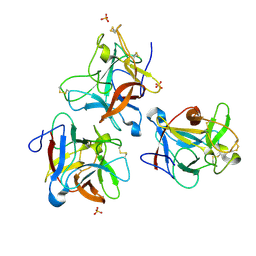 | |
8WIN
 
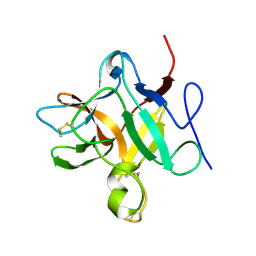 | |
8WE5
 
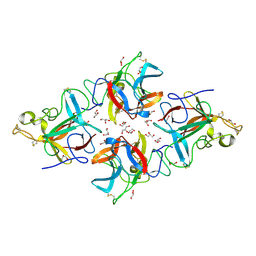 | |
8WFO
 
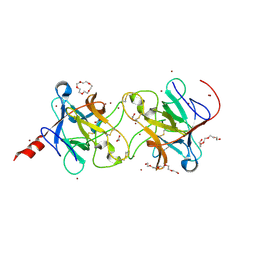 | |
8WI1
 
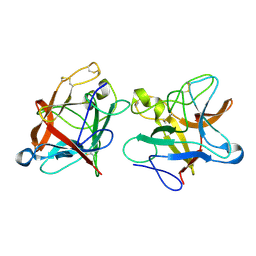 | |
8WK1
 
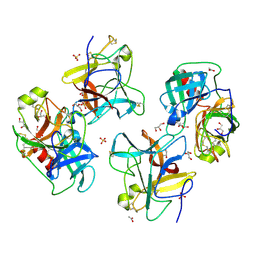 | |
8WKB
 
 | |
5XDR
 
 | | Crystal structure of human DEAH-box RNA helicase DHX15 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15, ... | | Authors: | Murakami, K, Nakano, K, Shimizu, T, Ohto, U. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7DJR
 
 | | Crystal structure of SARS-CoV-2 main protease (no ligand) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Deetanya, P, Wangkanont, K. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Interaction of 8-anilinonaphthalene-1-sulfonate with SARS-CoV-2 main protease and its application as a fluorescent probe for inhibitor identification.
Comput Struct Biotechnol J, 19, 2021
|
|
3WR7
 
 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
6M4F
 
 | | Crystal structure of the E496A mutant of HsBglA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase-like enzyme, ... | | Authors: | Uehara, R, Iwamoto, R, Aoki, S, Yoshizawa, T, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a GH1 beta-glucosidase from Hamamotoa singularis.
Protein Sci., 29, 2020
|
|
