3OFE
 
 | |
3OFH
 
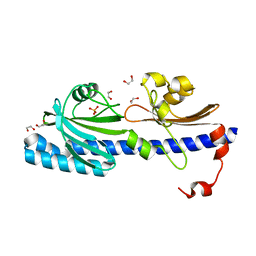
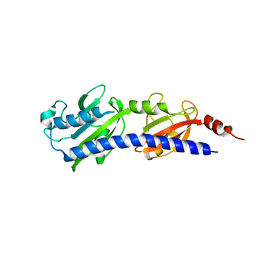
 | | Structured Domain of Mus musculus Mesd | | Descriptor: | LDLR chaperone MESD | | Authors: | Collins, M.N, Hendrickson, W.A. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural Characterization of the Boca/Mesd Maturation Factors for LDL-Receptor-Type beta-Propeller Domains
Structure, 2011
|
|
3PRA
 
 | |
3LIF
 
 | |
3LIE
 
 | |
3LIC
 
 | |
3LCK
 
 | |
3LI9
 
 | |
3LI8
 
 | |
3LIA
 
 | |
3M74
 
 | |
3M7E
 
 | |
3M7L
 
 | |
3M77
 
 | |
3LID
 
 | |
3LIB
 
 | |
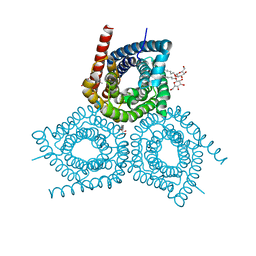
3M7C
 
 | |
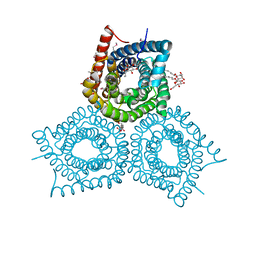
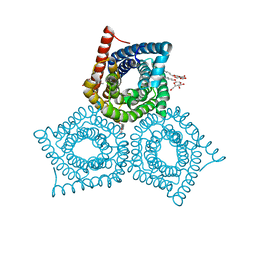
3M71
 
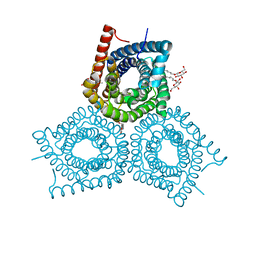 | | Crystal Structure of Plant SLAC1 homolog TehA | | Descriptor: | Tellurite resistance protein tehA homolog, octyl beta-D-glucopyranoside | | Authors: | Chen, Y.-H, Hu, L, Punta, M, Bruni, R, Hillerich, B, Kloss, B, Rost, B, Love, J, Siegelbaum, S.A, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-03-16 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Homologue structure of the SLAC1 anion channel for closing stomata in leaves.
Nature, 467, 2010
|
|
3M72
 
 | |
3M78
 
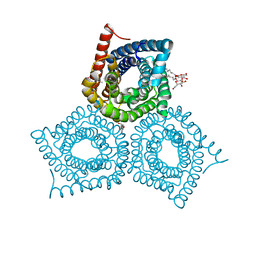 | |
3M70
 
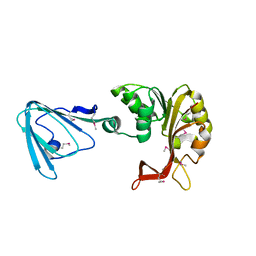 | |
3M75
 
 | |
3M76
 
 | |
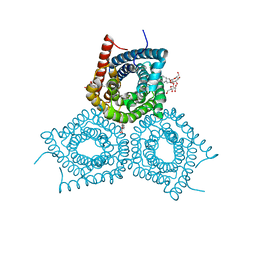
3M73
 
 | |
3M7B
 
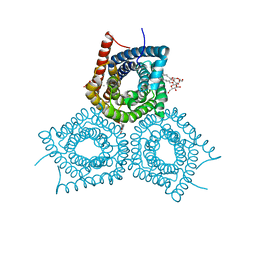 | |
